List of antibacterial drugs
| S. No. | Structure | Drug | Mechanism of action | Mechanism of resistance | References |
|---|---|---|---|---|---|
| 1 | 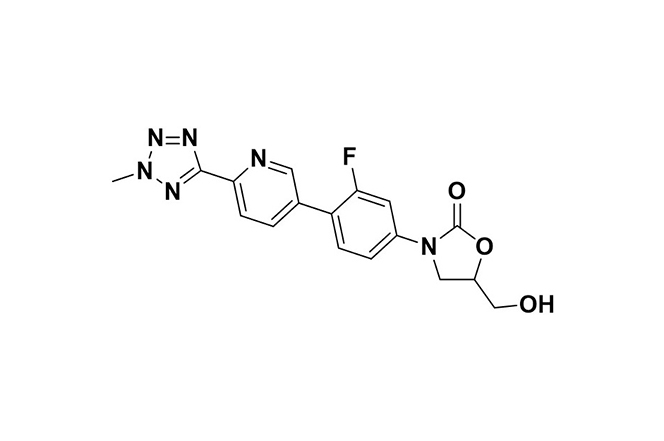 | Tedizolid | Binds to 23S rRNA in the 50S ribosomal subunit, blocking protein synthesis initiation and aminoacyl-tRNA binding in bacteria. | Resistance arises through 23S rRNA mutations or cfr gene transfer, reducing the drug’s ribosomal binding affinity. | [96] |
| 2 | 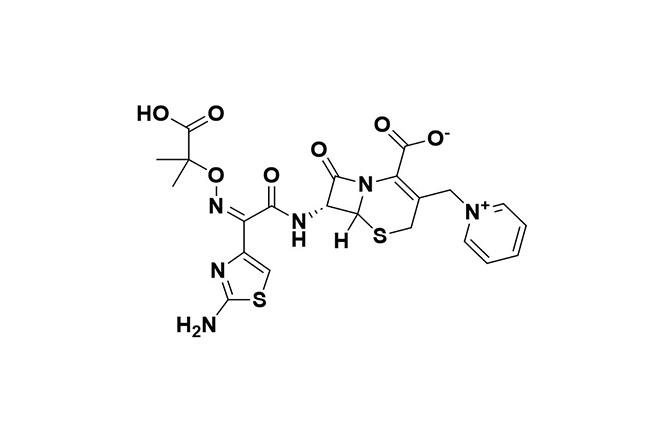 | Ceftolozane-tazobactam | Combines bactericidal effects of ceftolozane with tazobactam’s β-lactamase inhibition. | Resistance involves metallo-β-lactamases and AmpC hyperproduction, reducing drug efficacy. | [97] |
| 3 | 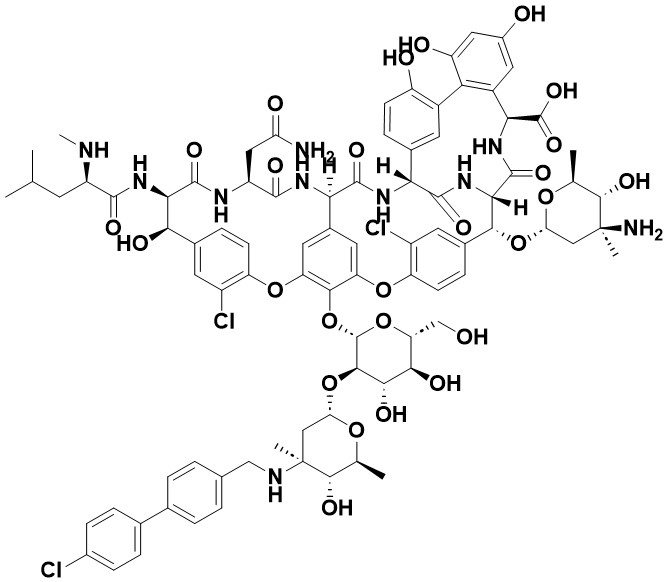 | Oritavancin | Inhibits peptidoglycan synthesis by binding to D-Ala-D-Ala and D-Ala-D-Lac, blocking transglycosylation and transpeptidation. | Resistance due to changes in VanB operon expression or D-Lac precursor presence, which reduce drug sensitivity. | [98] |
| 4 | 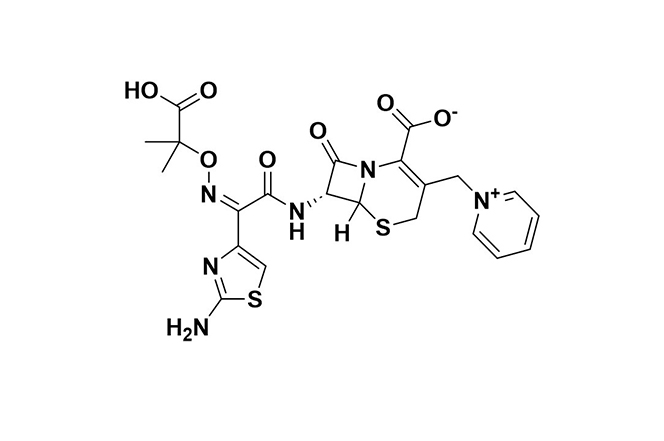 | Ceftazidime-Avibactam | Ceftazidime disrupts cell wall synthesis via penicillin-binding proteins (PBPs); avibactam inhibits ceftazidime breakdown by β-lactamases. | Resistance stems from metallo-β-lactamases, specific class D enzymes, mutations in β-lactamase enzymes (e.g., KPC-3), along with reduced permeability and efflux. | [99] |
| 5 | 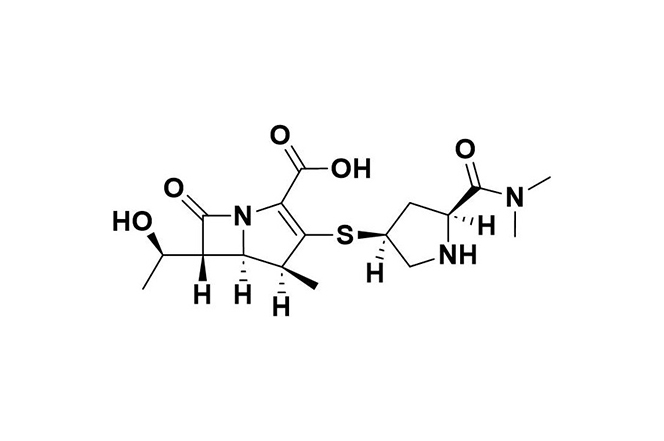 | Meropenem | Inhibits cell wall synthesis by binding to PBPs; vaborbactam enhances efficacy by inhibiting class A β-lactamases. | Resistance mechanisms include class B and D carbapenemases, altered PBPs, and increased efflux pump activity. | [100] |
| 6 | 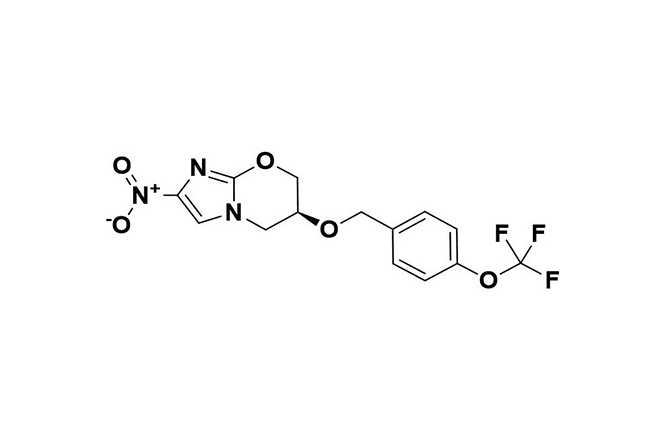 | Pretomanid | In anaerobic conditions, generates reactive intermediates that inhibit respiration in non-replicating bacteria and disrupt protein and lipid synthesis in replicating bacteria. | Resistance primarily occurs through mutations in activation-related genes (e.g., Ddn, Fgd, fbiA/B/C), sometimes affecting CofC or rplC and conferring dual resistance with linezolid. | [101] |
| 7 | 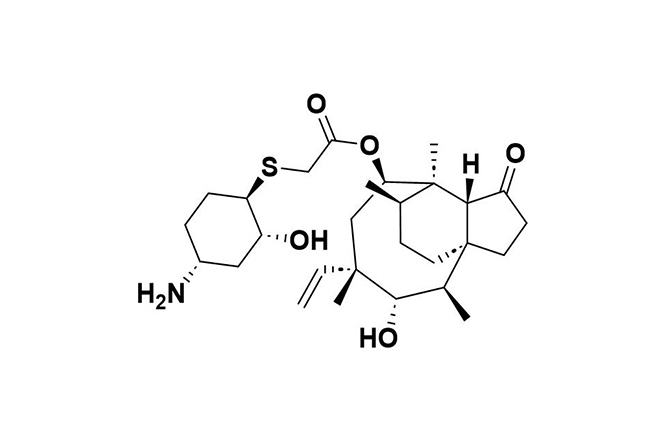 | Lefamulin | Binds to the 50S ribosomal subunit, inhibiting peptide bond formation and protein synthesis in bacteria. | Resistance arises from genes like vga(A), cfr, and mutations in 23S rRNA and ribosomal proteins (rplC, rplD). | [102] |
| 8 | 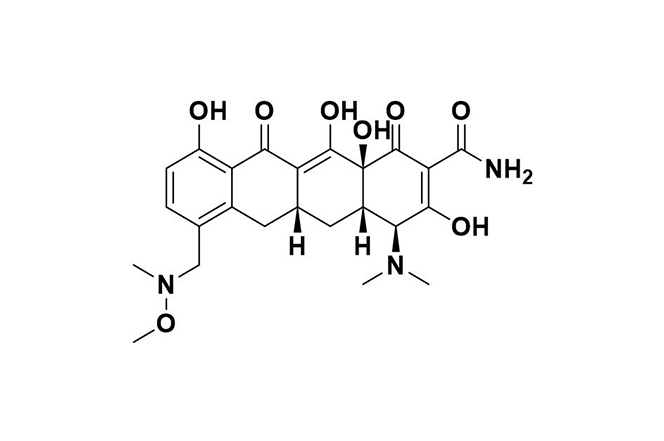 | Sarecycline | Binds to the bacterial ribosome’s A site, blocking mRNA translation; also reduces inflammation by suppressing cytokines and neutrophil activation. | Resistance occurs through degradation, rRNA mutations, ribosomal protection (e.g., TetM, TetO), and efflux pumps like TetA. | [103] |
| 9 | 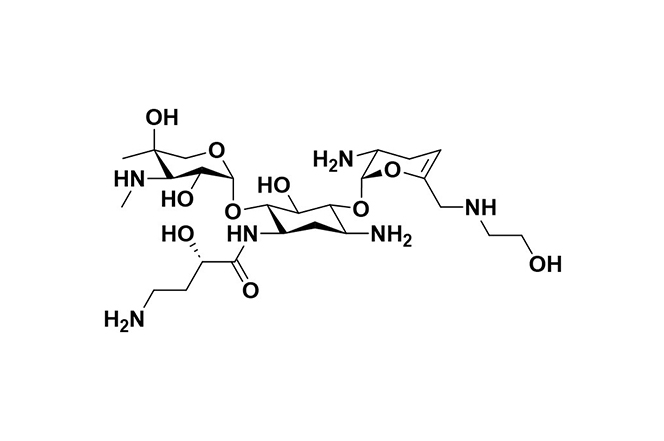 | Plazomicin | Inhibits protein synthesis by binding to the A-site of 16S rRNA in the 30S ribosomal subunit in Gram-negative bacteria. | Resistance is mostly due to aminoglycoside-modifying enzymes (AMEs), target site alterations, and changes in porin or efflux pump expression. | [104] |
| 10 | 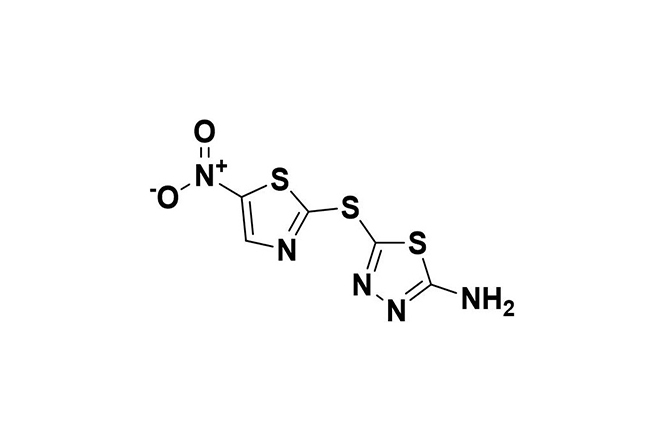 | Halicin | Exhibits broad-spectrum antibacterial action by damaging DNA and disrupting the proton motive force (PMF), impeding bacterial adaptation and resistance. | Resistance is rare and mainly involves mutations affecting protein synthesis, transport, and nitroreduction. | [105] |
| 11 | 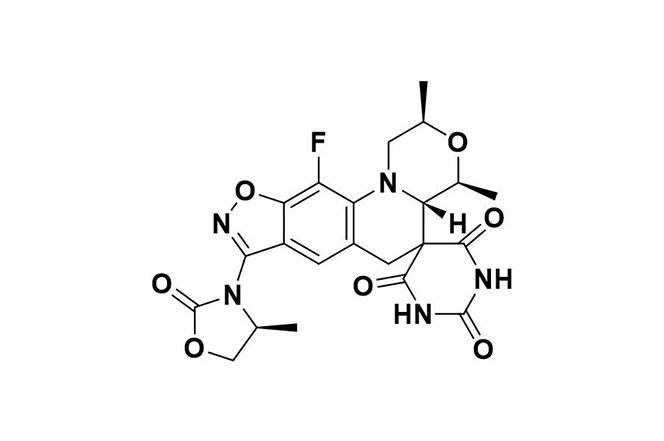 | Zoliflodacin | Inhibits bacterial type 2 topoisomerase, primarily targeting DNA gyrase, preventing fluoroquinolone cross-resistance and double-strand DNA breaks. | Resistance is uncommon, associated with gyrB gene alterations, and presents minimal cross-resistance with other antibiotics. | [106] |
| 12 | 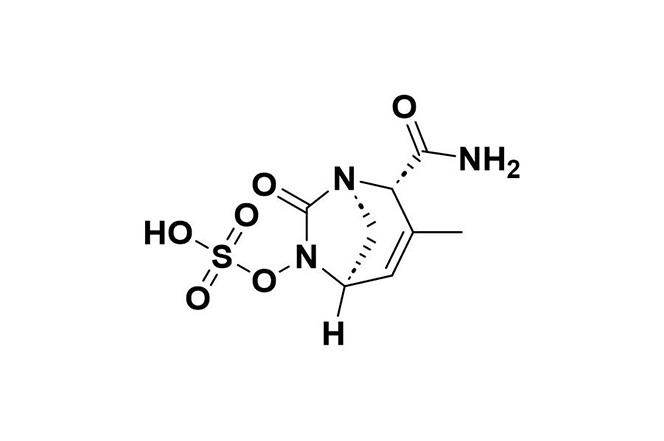 | Durlobactam | Restores Acinetobacter baumannii’s susceptibility to sulbactam by inhibiting class D carbapenemases, β-lactamases, and reducing PBP2. | Resistance may occur through metallo-β-lactamases, certain PBP mutations, and the AdeIJK efflux system. | [107] |
| 13 | 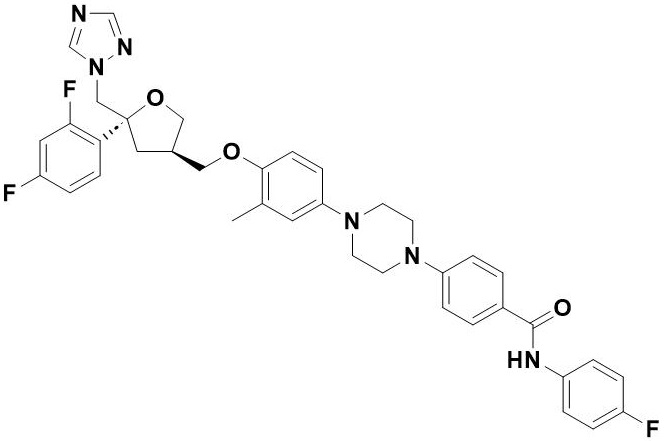 | Opelconazole | Inhibits 14-α-demethylase (CYP51A), blocking ergosterol formation in fungal cell membranes, effective against Aspergillus and Mucorales. | Resistance primarily arises from ERG11 gene mutations, notably TR34/L98H in cyp51A, which reduce drug binding efficacy. | [108] |