Affiliation:
1Izmir Biomedicine and Genome Center, 35340 Izmir, Turkey
2Department of Medical Biology, Faculty of Medicine, Dokuz Eylul University, 35340 Izmir, Turkey
Email: gulcin.cakan@ibg.edu.tr
ORCID: https://orcid.org/0000-0002-6356-5979
Affiliation:
3Department of Molecular Biology and Genetics, Ihsan Dogramaci Bilkent University, 06800 Ankara, Turkey
ORCID: https://orcid.org/0009-0006-1393-0620
Affiliation:
3Department of Molecular Biology and Genetics, Ihsan Dogramaci Bilkent University, 06800 Ankara, Turkey
ORCID: https://orcid.org/0009-0003-0635-9495
Affiliation:
1Izmir Biomedicine and Genome Center, 35340 Izmir, Turkey
ORCID: https://orcid.org/0000-0003-3822-9073
Affiliation:
3Department of Molecular Biology and Genetics, Ihsan Dogramaci Bilkent University, 06800 Ankara, Turkey
Email: konu@fen.bilkent.edu.tr
ORCID: https://orcid.org/0000-0002-6223-5329
Explor Dig Dis. 2023;2:44–55 DOl: https://doi.org/10.37349/edd.2023.00017
Received: December 29, 2022 Accepted: February 13, 2023 Published: April 26, 2023
Academic Editor: Maria L. Martinez-Chantar, University of Deusto, Spain
The article belongs to the special issue Drug-induced Liver Injury: From Bench to Clinical Application
Zebrafish as a preclinical drug induced liver injury (DILI) model provides multiple advantages ranging from ease of breeding and maintenance, availability of different strains and transgenic fish amenable to study liver function, and highly conserved liver structure and function with the human liver. In this review, the authors have aimed to provide an account of the metabolic enzymes that take roles in drug detoxification in both human and zebrafish in a comparative manner and exemplify several recent models in studying liver functionality. Moreover, the authors emphasize the difficulties associated with studying idiosyncratic DILI in preclinical models and propose that zebrafish could be an important complement to mice in testing functions of genes that are associated with DILI with respect to different drugs in human genome-wide association studies (GWAS) Catalog. Finally, this review highlights the state-of-the-art in the development of novel transgenic reporter strains that can be used to study degree and molecular mechanisms of hepatotoxicity caused by drugs in zebrafish. All of these will help researchers to use effectively the available resources in the zebrafish DILI models, while advocating potential leads that can be taken to provide advancements in a better understanding and treatment of DILI.
Zebrafish is a genetically tractable, vertebrate model that has been proven efficient in disease modeling, drug efficacy, and toxicity evaluation [1, 2]. Fast embryonic development and accessibility of embryos and larvae for imaging and drug treatment are leading advantages. Zebrafish reproduce via external fertilization, and the embryo is accessible from time 0 for imaging and drug treatments delivered in rearing water. The protective chorion is a selective membrane that prevents penetration of drugs, however, once the larvae hatch from the chorion at the end of day 2, it is fully accessible to drugs (Figure 1). Zebrafish embryo and early larvae [3–5 days post fertilization (dpf)] get their nutrition from the yolk, which decreases in volume gradually while the internal organs such as gut and liver develop (Figure 1). Zebrafish liver is detectable as of 3 dpf (Figure 1), but it becomes fully functional when the larvae reach the 5 dpf stage [3]. Differentiated hepatocytes are first detected at 44–48 hours post fertilization (hpf) followed by differentiation of biliary cells between 50–70 hpf and vascularization onset at 55–60 hpf. Finally hepatic stellate cells are detected by 96 hpf and liver outgrowth is completed between 96–120 hpf [4–6].
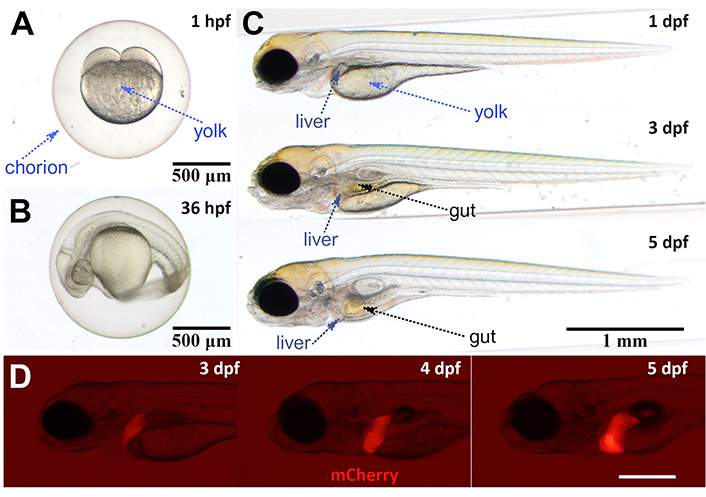
Developmental stages of zebrafish and liver. A. Two-cell stage zebrafish embryo is visible at 1 hpf. The chorion provides protection to the embryo while the yolk supplies all the required nutrients; B. the embryo develops in the chorion until end of day 2. Thirty-six hpf embryo is still in the chorion; C. larval stages begin after hatching from the chorion. Liver can be seen as a small tissue at 3 dpf while gut is visible as of 4 dpf; D. transgenic reporter line fabp10a:mCherry is used to label hepatocytes. Liver tissue is visible as of 3 dpf with this line. Scale bar: 500 µm. Images A–C are recorded with Olympus SZX10 Stereomicroscope and SC50 camera, and image D is recorded with Olympus SZX16 Fluorescent Stereomicroscope and XC50 camera. fabp10a: fatty acid binding protein 10a (promoter); mCherry: monomeric Cherry
Hepatocytes, stellate cells, biliary epithelial cells, and bile ducts are present in the zebrafish liver although their structural organization is different than that of human [3, 4]. Liver macrophages are also present in both the larval and adult liver tissues, which are discussed comprehensively in a recent review [7]. The differences between the structural architectures of the zebrafish and human liver are discussed and illustrated in two previous reviews that describe a more scattered appearance of portal vein, hepatic artery, central vein, and bile ducts in the zebrafish liver [8, 9]. Moreover, in the zebrafish liver, the hepatocytes are arranged in tubules rather than in plates. In adult zebrafish, the male and female liver structures and metabolism differ significantly, which is a factor to be considered in drug induced liver injury (DILI) studies [9]. On the other hand, larval stages of 2–5 dpf as well as 5–10 dpf provide attractive alternatives for DILI studies.
In this review, we opted to i) cover the current knowledge on major drug metabolizing enzymes in the zebrafish liver; ii) provide examples to the discovery and validation of genome-wide association studies (GWAS) genes/variants in zebrafish models with a focus on liver disease and idiosyncratic DILI; iii) and highlight the state-of-the art zebrafish technology that can facilitate DILI studies.
The liver detoxification system has important distinct phases that include membrane transportation (initial phase), oxidation reduction reactions (phase I), enzymatic based conjugation (phase II), and efflux of metabolites (phase III) [10]. Zebrafish liver is a valuable model for drug metabolism studies since the major enzymes of drug metabolism are expressed and functional in zebrafish larval and adult liver. Phase I of drug metabolism is driven by cytochrome p450 (CYP) enzymes that induce oxidation, reduction, and hydrolysis of xenobiotics and toxins. Among the 57 CYPs in human genome, 15 isoforms in CYP1–3 families are responsible for oxidation of most xenobiotics [11]. A systematic analysis of idiosyncratic DILI cases with Roussel Uclaf Causality Assessment Method (RUCAM) showed that most drugs are metabolized by CYP isoforms of which CYP3A4/5 is used close to 50% of the time, followed by CYP2C9, CYP2E1, CYP2C19, CYP1A2, and CYP2D6 [12]. There are 94 -cyps in the zebrafish genome representing all 18 families that are present in the human genome [13]. The enzymes that regulate xenobiotic metabolism falling into Cyp1–3 families are more complex and diverse in zebrafish having multiple subfamilies, which are discussed elsewhere in detail [13–15]. Zebrafish orthologs of -cyp1–3 family genes and their functions in human liver are summarized in Table 1. Of note, the zebrafish cyp1A shows similarity to human CYP1A1 and A2 genes, and the zebrafish cyp1B1 and human CYP1B1 share a very similar gene structure. CYP1A and CYP1B subfamilies are known to be important in metabolism of steroids and various drugs in vertebrates. Complexity increases in the cyp2 gene family which contains 47 members in the zebrafish genome. Goldstone et al. [13] proposed that tandem localization of cyp2y, cyp2y3, and cyp2y4 resembles the cluster of drug metabolizing CYP2A6, CYP2A13, CYP2B6, CYP2F1, and CYP2S1 in the human genome. Zebrafish cyp3 family, on the other hand, consists of cyp3a65 and cyp3c1–3c4, among which cyp3a65 shows 54% sequence identity to human CYP3A4. cyp3a65 expression is detected in the liver and intestines of adults and larvae, which was shown to be further increased upon xenobiotic treatment [14, 16].
Zebrafish genes of xenobiotic metabolizing CYPs in cyp1–3 families. Drug detoxifying CYPs mentioned in the text are listed in the table. Zebrafish gene and corresponding human gene are listed in the same row. Functional conservation needs to be tested
| Gene family | Zebrafish gene | Human gene | Function |
| CYP1 | cyp1a, cyp1b1, cyp1c1,2, cyp1d1 | CYP1A1,2, CYP1B1, CYP1D1P | Catalyzing oxidative biotransformation of xenobiotics, dioxin-induced cardiovascular toxicity [14] |
| CYP2 | cyp2r1 | CYP2R1 | Microsomal vitamin D 25-hydroxylase [13] |
| - | cyp2u1 | CYP2U1 | Catalyzing ω and ω-1 hydroxylation of fatty acids [13] |
| - | cyp2n13, cyp2ps, cyp2v1, cyp2ad2,3,6 | CYP2J2 | Arachidonic acid epoxygenase [13] |
| - | cyp2ks | CYP2W1 | It is a tumor-specific CYP that oxidizes indole and chlorzoxazone, but not fatty acids [13] |
| - | cyp2y3,4 | CYP2A/B/F/S | Drug-metabolizing enzymes |
| CYP3 | cyp3a65 | CYP3A4 | Metabolism of various drugs |
| - | cyp3c1–4 | CYP3A3,4,7 | Mediates the formation of 5-hydroxydiclofenac (5-OHDF) and 6β-hydroxytestosterone [15] |
| CYP4 | cyp4f43 | CYP4F | Omega-hydroxylases of C16–C26 fatty acids [13] |
| - | cyp4v7,8 | CYP4V2 | - |
-: none
Further studies are required in order to better understand functions, substrate specificity, and regulations of the zebrafish CYPs. With advancement of the field, zebrafish models can help understanding of mechanisms of CYP involvement in DILI. The ease of transgenic zebrafish generation provides easy means of humanizing the zebrafish liver. For example, the expression of human CYP3A4 (hCYP3A4) and mCherry fusion in zebrafish hepatocytes via liver specific fabp10a promoter in the Tg(fabp10a:hCYP3A4-mCherry) transgenic line enhanced the metabolism of CYP3A4 substrates and this could be inhibited with an CYP3A4 inhibitor, ketoconazole [17].
Phase II drug metabolism and detoxification enzymes have zebrafish orthologs, and conserved functions of several enzymes have been shown. Here, the recent knowledge on zebrafish UDP-glucuronosyltransferases (UGTs) and glutathione-S-transferases (GSTs) is discussed. Cytosolic sulfotransferases (SULTs) form another major detoxifying enzyme family that has been studied extensively in zebrafish, and these are reviewed elsewhere [18–21].
Zebrafish -ugt gene family consists of 40 genes and 5 pseudogenes, which are diversified by presence of ugt1, 2 and 5 genes and several alternatively spliced forms [22]. Wang et al. [23] studied tissue specific expressions of the 40 zebrafish ugts and reported catalytic activities and substrate specificities of these zebrafish Ugts. gsts are expressed in the developing zebrafish embryo, larvae, and adult [24, 25]. Several zebrafish gsts were cloned and showed glutathione (GSH) conjugation activity in vitro when recombinantly produced [25]. A protein expression study showed that some Gst proteins were expressed as of 4 hpf, while others that belong to zeta, theta, and omega classes were shown to be expressed as of 72 hpf [24]. Expression of these proteins [Gsta1-2-3, glutathione-S-transferase theta 1a (Gstt1a), Gstz1-2-3, Gstm3, and Gsto1] increased steadily from 72 hpf to 168 hpf, while in adults only Gsta1-2-3, Gstt1a, and Gst1, 2, 3 proteins were found to be liver enriched suggesting these might be the main Gsts of phase II detoxification in zebrafish liver maturation [24].
Genome duplication in zebrafish is a complicating factor for some enzyme groups such as UGTs. Therefore, enzyme activity analyses are key to determining functional conservation. Moreover, the humanization approach that was used for CYP3A4 could be extended for other key enzymes to generate specific reporter lines.
Use of preclinical animal models is essential for validation of the emerging DILI biomarkers and/or variants from genome-wide screens [26–28]. Zebrafish model is well suited for such validation studies; and offers a plethora of assays and tools enabling preclinical DILI studies of different scales, ranging from small-to-large, in the intact organism.
Direct and indirect DILI, which are dose-dependently caused by agents inherently or indirectly toxic to the liver, respectively, are relatively easier to reproduce in model organisms than idiosyncratic hepatotoxicity, which is largely due to the individual’s metabolic or immune response [29, 30]. A recent review has further highlighted potential difficulties in aligning successfully the human and other animal models of idiosyncratic DILI, including that of zebrafish [31]. The ability to apply in vivo genetic and/or environmental modulations, such as challenging animals with inflammatory agents in rodent models, could lead to induction of DILI at therapeutic doses, as reviewed by Segovia-Zafra et al. [32].
In humans, GWAS is the main-stream analysis platform for polygenic traits like idiosyncratic DILI, for which the effect size of a polymorphism, e.g., single nucleotide polymorphism (SNP), is often small while a large number of loci could be associated with the trait [33]. Zebrafish has been used as a model to demonstrate causality of the prioritized variants from GWAS screens in the observed phenotype. However, many novel variants are intronic and/or not conserved; accordingly, not the variant itself but the overexpression or knockout/knockdown models of the gene are used in validation studies. For instance, rs8004664, an intronic SNP found in forkhead box N3 (FOXN3), has been associated with an increase in fasting glucose levels in humans, and the researchers generated transgenic zebrafish overexpressing the human or zebrafish gene and showed that overexpression resulted in excess glucose production in both models [34]. Similarly, different non-coding variants of ADP-ribosylation factor 6 (arf6) have been associated with biliary atresia and the arf6 knockdown experiments performed in zebrafish resulted in intrahepatic biliary network sparsity and decreases in bile secretion supporting arf6 as a potential susceptibility locus in biliary disease [35]. With respect to DILI, a workflow has been proposed to narrow down candidates obtained from the human GWAS based on the presence of a significant zebrafish-human orthology, expression in the liver, and disease association [36]. Accordingly, the authors validated changes in expression via morpholino knockdown of selected zebrafish genes that affected hepatocyte function and/or liver development. Recent studies employ clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein 9 (Cas9) mediated genome editing technology to validate orthologous gene function via generating various allele modifications that range from nonsynonymous to synonymous in zebrafish and screen the relevant phenotype in embryos or larvae. One example comes from understanding genetic factors affecting cardiac rhythm [37]. A recent review on variant discovery and validation of osteoporosis also suggests that G0 screening or F0 crispants can aid in rapid identification of candidate variants in zebrafish and hence speed up the discovery process [38]. Although not yet in DILI, a putative causal variant leading to a null phenotype could be demonstrated to replicate the disease phenotype in zebrafish, for example in cleft palate [39].
There are many GWAS carried out to link DILI to human genetic traits in the last decades, and online tools such as GWAS Catalog provide searchable databases of keywords of genes and diseases [33, 40]. Indeed, in GWAS Catalog, there is a specific category for DILI related studies and associations (i.e., EFO_id: EFO_0004228) while other liver associated phenotypes including hepatotoxicity due to other causes also exist. EFO_0004228 contains multiple GWAS datasets; a few of which have large sample sizes containing more than 10,000 cases and controls in total, while most datasets are of medium to small size. In GWAS Catalog, DILI studies also comprise a range of drugs that include general antibiotics (e.g., flucloxacillin), anti-tuberculosis drugs (e.g., rifampicin), and more specific drugs targeting genes and pathways, such as lapatinib and interferon-beta (IFN-β), and hence presenting an excellent tool for datamining. Accordingly, close to 60 human genes can be found in EFO_0004228 category of GWAS Catalog by December, 2022. However, it is still a challenge to decide on which human gene and/or genomic variant to pursue specifically in zebrafish although several databases and tools are available that can assist in the decision process.
The teleost lineage, to which zebrafish belongs, has gone through an extra round of whole genome duplication thus, the zebrafish genome may have duplicated copies of human disease genes [41, 42]. The zebrafish orthologs of selected human loci can be gathered from BioMart database in Ensembl via the setting the “orthology type” filter, to “one-to-one” and “one-to-many” and hence, filtering out potentially ambiguous hits that may originate from using “many-to-many” type genes [42, 43]. BioMart also allows the researchers to specify sequence features, e.g., the zebrafish and human Ensembl IDs, and chromosomal locations. Similarly, use of ribosome-upstream open reading frames (Ribo-uORFs), which incorporates zebrafish genomic data, can be further useful for identification of open reading frames in 5’ untranslated region (5’UTR) and their associated expression quantitative trait loci (eQTL) analysis [44]. Other additional information on the liver-specific or liver-enriched expression of the prioritized genes or the gene’s presence in specific hepatocyte cell clusters can be of further use as demonstrated before [36]. Specialized expression databases are also available, such as a webserver called Cell Landscape, in which cell/tissue specific single cell RNA-seq (scRNA-seq) profiles are provided for human, mouse, and zebrafish for a given transcript [45].
In the context of GWAS, zebrafish is a promising vertebrate model for validation of candidate variants or genes, found via GWAS, by examining their effect on the model’s liver phenotype [36]. Herein, we provide a table for potential advantages and disadvantages of zebrafish in validating GWAS loci from idiosyncratic DILI studies (Table 2).
The advantages and disadvantages of using zebrafish for validation of GWAS loci from DILI studies
| Advantages | Disadvantages |
|---|---|
|
|
|
|
|
|
|
|
|
|
|
|
Moreover, the polygenic analysis of genome-wide variant landscape, and not the analysis of only a single or few variants, is more likely to advance our understanding of DILI. One such example comes from the case-control cohort study of DILI, driven by TAK-875 (fasiglifam), a drug withdrawn while going through phase III clinical trials against diabetes 2 due to rare events of severe acute hepatocyte injury [46, 47]. Later, Mosedale et al. [48] showed the importance of using preclinical models for the polygenic analysis in TAK-875 driven liver injury via high throughput expression profiling and eQTL analyses, using the 45 collaborative cross (CC) mouse lines, a genetically diverse collection of inbred strains. The authors have identified mitochondrial dysfunction and oxidative stress related pathways as being the most strain dependent, and hence characterized presence of genetically driven transcriptomic changes while identifying candidate eQTLs for further study [48]. A similar approach can be taken for the zebrafish DILI models since there are thousands of zebrafish mutant and genetically modified strains that can be controllably bred to generate a collaborative collection of strains for identifying polygenic structure of DILI and other complex diseases. With the discovery of novel and population specific hepatocyte eQTLs in humans, preclinical zebrafish GWAS screens in well-designed drug exposure studies could have a significant impact on discovery and validation of novel genotype-phenotype associations in idiosyncratic DILI [49].
On a final note, Teschke and Danan [30], upon reviewing thousands of idiosyncratic DILI registries, provide a list of the drugs most frequently resulted in idiosyncratic DILI, e.g., amoxicillin-clavulanate, flucloxacillin, and atorvastatin. Such drugs if tested at therapeutic doses in select zebrafish mutants with or without exposure to additional environmental DILI risk factors may prove fruitful for improving our understanding of idiosyncratic DILI. The causality between the drug and its associated hepatoxicity also needs to be robustly assessed, using methods such as RUCAM, to accurately distinguish between the intrinsic and idiosyncratic DILI cases [30]. Moreover, similar methodologies need to be adapted to the zebrafish idiosyncratic DILI models; however, the strong dependency of RUCAM on the levels as well as ratio of the liver enzymes might make this difficult in embryos and larvae. Due to the small size of the larval and even adult zebrafish monitoring, these enzymes from sera could not be feasible. New methodologies amenable for accurate and continuous live detection of existing and new biomarkers are needed for developing effective zebrafish idiosyncratic DILI models.
Embryonic and early larval stages of zebrafish provide advantages of easy imaging and amenability to medium-high throughput screening. Identification of the liver specific fabp10a, and its promoter sequence allowed successful development of liver reporter lines that drive the reporter gene expression in hepatocytes [50, 51]. Assessment of the changes in liver size upon drug exposure is inarguably the most widely used tool for screening drug related injury in zebrafish models [52–56]. Moreover, lineage specific reporter lines marking biliary cells, e.g., Tg(Tp1glob:EGFP) and Tg(krt18:EGFP), and stellate cells, e.g., Tg(hand2:EGFP), are available to assess the effects of DILI on liver cells [4, 57, 58]. Another interesting approach is to use translating ribosome affinity purification (TRAP), an effective methodology that was applied to profile hepatocyte or biliary cell specific transcripts; in transgenic lines Tg(krt18:TRAP) and Tg(lfabp:TRAP) that drives TRAP expression under keratin-18 promoter and fabp10a promoters, respectively [58]. These TRAP lines can further be used to profile DILI response upon exposure to xenobiotics.
Another approach is to search for candidate biomarkers for DILI using high-throughput omics studies and meta-analysis [59–61]. For instance, Poon et al. [61] conducted microarray analysis on livers of zebrafish in which DILI was induced by three different hepatotoxin drugs, namely amiodarone, diclofenac, or flutamide, and discovered candidate biomarker genes that were upregulated in all. Furthermore, the analysis of spatiotemporal expression and induction of these common genes have led to identification of cyp2n13, cyp2k18, gstt1b, and glutathione reductase (gsr) as potent biomarker genes in zebrafish DILI in both the larval and adult stages. Enhanced green fluorescent protein (EGFP) reporter lines that were generated by promoter cloning or green fluorescent protein (GFP) tagging of the endogenous gene via FOSMID recombination eventually led to the development of cyp2n13, cyp2k18, gstt1b, and gsr reporter lines as potential tools for high throughput screens for any given drug’s potency to induce DILI [61]. cyp2n13 and gsr are highly conserved between zebrafish and human, while cyp2k18 and gstt1b are orthologous to human CYP2W1 and GSTT1, respectively [13].
More recently, a hepatic apoptosis reporter line Tg(pck1:Casper3GR) was generated by Higuchi et al. [62], which may be used as another screening tool. Promoter sequence of phosphoenolpyruvate carboxy kinase 1 (pck1) gene, was used to drive expression of a Casper3GR, which is a dual fluorescent probe that is cleaved by caspase-3, hence provides a fluorescence resonance energy transfer (FRET)-based apoptosis sensor in the liver [62].
Expression of bacterial nitroreductase gene in hepatocytes is used for targeted ablation of liver cells via exposure to prodrug metronidazole [55]. Recently this tool has been used to identify hepatoprotective small molecules in a screen and BML-257, an AKT inhibitor, was found to provide protection against liver damage caused by acetaminophen (APAP) and isoniazid [63]. Moreover, according to a recent review, the role of ferroptosis in DILI, when compared to other liver diseases, remains relatively unexplored with most of the research being based on APAP induced DILI [64]. New evidence is emerging in support of the therapeutic potential of scavenging reactive oxygen species, e.g., peroxynitrite, release of H2S and ferroptosis inhibitors in ameliorating the liver damage induced by APAP in vitro and in mice models [65]. Interestingly, a novel sensor of peroxynitrite has also been applied to the zebrafish model which demonstrated its translational significance [66].
One of the concerns using the highly promising zebrafish larval models for DILI assessment could be that embryos and early larvae may not be fully functional in drug metabolism. Several studies provide evidence that zebrafish larvae can indeed serve as functional DILI models even at an early stage where the transparency of the larvae provides excellent opportunities for high-throughput screening. Embryonic zebrafish hepatocytes are detectable as early as 36 hpf, however hepatic differentiation continues until 96 hpf while the liver development is completed by 120 hpf which corresponds to 5 dpf resulting in a fully functional xenobiotic metabolism in addition to other liver functions [3]. Interestingly, the majority of DILI studies are conducted in zebrafish between the developmental stages spanning 48 hpf to 5 dpf. A recent study by Nawaji et al. [15] investigated the reported spatiotemporal expression of -cyp genes as well as their induction and activity upon exposure to certain substrates during the liver development stages and adult fish. This study revealed that several drug metabolizing cyps were expressed well before liver development was completed, in primordial intestine, primordial bladder, or primordial liver. Moreover, use of CYP inducers and substrates allowed researchers to demonstrate the metabolic activity of some zebrafish Cyp enzymes (Cyp1a2, Cyp2c9, and Cyp3a4/5) in these extrahepatic tissues before liver maturation [15]. Therefore, use of zebrafish exposed to xenobiotics at early stages may already activate the phase I of drug metabolism. On the other hand, direct comparison of 5 dpf and older larvae vs. younger embryos has not been reported.
Zebrafish liver can be used as a DILI model at larval and adult stages. Fast maturation, amenability to live imaging, and drug treatment combined with genetic tools make zebrafish a unique vertebrate model. Conservation of drug metabolism enzymes including CYPs and UGTs is reported in recent studies. Future studies are needed to better describe substrate specificity and functional conservation of these enzyme families. Zebrafish can also be invaluable for modeling idiosyncratic DILI in addition to intrinsic DILI by means of developing transgenic and knockout-knockin models of candidate loci discovered from GWAS that are ever increasing in number. These models are likely to allow for testing novel therapeutic strategies while increasing our understanding of the genotype-phenotype associations in DILI. Moreover, the wide array of developed reporter zebrafish lines opens the possibility to generate readouts for lineage specific or enzyme specific phenotypes upon exposure to drug candidate. In order to enable fast and reliable screening of large number of drug candidates, the continued efforts of zebrafish researchers are needed to expand the repertoire of DILI sensor reporter lines, and describe each line’s potency in reflecting different drug metabolism processes.
APAP: acetaminophen
arf6: ADP-ribosylation factor 6
CYP: cytochrome p450
DILI: drug induced liver injury
dpf: days post fertilization
eQTL: expression quantitative trait loci
fabp10a: fatty acid binding protein 10a
gsr: glutathione reductase
GSTs: glutathione-S-transferases
Gstt1a: glutathione-S-transferase theta 1a
GWAS: genome-wide association studies
hpf: hours post fertilization
mCherry: monomeric Cherry
RUCAM: Roussel Uclaf Causality Assessment Method
TRAP: translating ribosome affinity purification
UGTs: UDP-glucuronosyltransferases
Authors thank Emine Gelinci and Ebru Önal for recording zebrafish images.
GCA: Conceptualization, Visualization, Writing—review & editing. AMA, MCC: Visualization, Writing—review & editing. KAA: Writing—review & editing. OK: Conceptualization, Writing—review & editing. All authors read and approved the submitted version.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
GCA and OK are supported by The Scientific and Technological Research Council of Türkiye (TUBITAK). GCA: [119S698], [118C484], OK: [116Z388]. GCA and OK are supported by the European Union COST action [CA17112] PRO-EURO-DILI-NET and Horizon Europe [101095679] project. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
© The Author(s) 2023.
Copyright: © The Author(s) 2023. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Alejandro Cueto-Sánchez ... Marina Villanueva-Paz
Hanghang Wu ... Francisco Javier Cubero
Qian Wei ... Jinsheng Guo
Miren García-Cortés ... Alberto García-García
