Abstract
Aim:
Modifications in long non-coding RNA (lncRNA) expression are associated with inflammation and fibrosis in chronic liver diseases. It has been recently demonstrated that human liver stem cells (HLSCs) and their extracellular vesicles (EVs) can effectively reduce inflammation and fibrosis in a murine model of non-alcoholic steatohepatitis (NASH). Now it has been evaluated whether EVs can modify the expression of inflammation-related lncRNAs in NASH liver.
Methods:
To induce NASH, severe combined immunodeficient mice were fed with a methionine-choline-deficient diet for 4 weeks. After 2 weeks of diet, 2.5 × 109 EVs were intravenously injected twice a week. An array of 84 inflammation-related lncRNAs was performed on the RNA isolated from NASH livers, and the expression of 14 selected lncRNAs was then validated by real-time polymerase chain reaction (PCR) analysis. Expression levels of maternally expressed gene 3 (Meg3) were further evaluated in vitro, in an activated human hepatic immortalized stellate cell line (LX-2) stimulated with EVs.
Results:
The screening showed an altered lncRNA expression profile in the liver of NASH mice, in respect to control healthy mice. EV treatment modulated several inflammation-related lncRNAs in NASH livers. Real-time PCR validation of array results indicated that EVs restored to normal levels the expression of 10 lncRNAs altered in NASH. In particular, EV stimulation reduced Meg3 expression levels, which were increased in NASH as well as in activated LX-2.
Conclusions:
HLSC-EVs regulate the expression of inflammation-related lncRNAs impaired in NASH livers and in an in vitro model of liver fibrosis.
Keywords
Non-alcoholic fatty liver disease, non-alcoholic steatohepatitis, chronic liver disease, human liver stem cells, extracellular vesicles, long non-coding RNAsIntroduction
Non-alcoholic fatty liver disease (NAFLD) is typically characterized by an abnormal increase of fat in the liver and is now considered one of the most common causes of chronic liver disease worldwide. Unfortunately, about 30% of patients with NAFLD develop an aggressive form of liver steatosis, known as non-alcoholic steatohepatitis (NASH), characterized by inflammation and fibrosis that may progress to cirrhosis, hepatocellular carcinoma, and other liver-related complications. In Western country populations with NAFLD, the prevalence of NASH is 20–30% and this percentage is steadily increasing, along with the augmented prevalence of insulin resistance, hypertension, and obesity [1–3]. At present, organ transplantation represents the standard therapy for NASH and other end-stage liver diseases. However, the availability of such treatment is limited by the high costs, the need for lifelong immunosuppressive agents, and the shortage of organ donors [2].
In the last decade, stem cell therapy has emerged as a potential alternative for the treatment of end-stage liver disease, including NASH. It has been recently demonstrated that human liver stem cells (HLSCs) and their extracellular vesicles (EVs) can ameliorate liver function and histology in a murine model of NASH [4, 5].
EVs are a non-homogenous population of membranous nanoparticles that include apoptotic bodies (1–5 μm in size), microvesicles or ectosomes (100–1,000 nm), and exosomes (50–150 nm). Emerging evidence indicates that, by transferring proteins, lipids, and various species of RNAs to recipient cells, EVs derived from stem/progenitor cells may induce epigenetic changes in injured target cells, thus triggering regenerative programs in the damaged tissue [6, 7]. At molecular level, treatment with HLSC-EVs in NASH mice has been shown to restore the expression of specific markers of fibrosis to baseline levels [5].
The major cellular drivers of liver fibrosis are hepatic stellate cells (HSCs). Upon chronic liver injury, these vitamin A-storing cells undergo an activation process and transform into myofibroblast-like cells, expressing alpha-smooth muscle actin (α-SMA). Furthermore, activated HSCs are primarily responsible for the production of extracellular matrix components, such as alpha-1 type I collagen (COL1A1), that accumulate in the liver and contribute to the progression of fibrosis [8]. In an in vitro model of liver fibrosis, HLSC-EVs administration has been shown to reduce the expression of pro-fibrotic genes in activated HSCs [9].
Besides modulating pro-fibrotic transcripts in activated HSCs and NASH mice, it may be possible that HLSC-EVs regulate the expression of other RNA species involved in fibrosis. The contribution of long non-coding RNAs (lncRNAs) in the onset and progression of pathological conditions, including liver fibrosis, is currently under intensive investigation [10]. LncRNAs are non-protein coding transcripts with more than 200 nucleotides that are predominantly transcribed by RNA polymerase II [11]. In the nucleus, lncRNAs can bind to proteins, RNA and DNA to alter chromatin structure and modulate the transcription of neighboring and distant genes, while in the cytoplasm they can affect RNA stability, splicing, and translation [12, 13]. A growing body of research has focused on maternally expressed gene 3 (Meg3), a lncRNA located at 14q32, which involvement in fibrogenesis has been described in various organs, such as the liver [10], kidney [14, 15], heart [16, 17] and lung [18, 19].
The aim of this study is to investigate if HLSC-EVs can modulate the hepatic lncRNA expression profile in a murine model of NASH. Furthermore, in activated HSCs, it has been examined whether lncRNA Meg3, whose expression is impaired by NASH, can be restored to normal levels by HLSC-EV treatment.
Materials and methods
Isolation and characterization of HLSC-EVs
HLSCs were generated from one donor liver belonging to the category of standard risk. Anemocyte International (Gerenzano, Italy) obtained the ethical approval and consent to participate of the donor to generate the master cell bank by the National Transplant Center in accordance with Italian legislation. HLSCs were cultured as previously described to generate a master cell bank [4, 5, 20]. The phenotypic characterization of HLSCs is provided by Table S1. For this study, cryopreserved cells in fetal bovine serum (Euroclone, Pero, Milano, Italy) in the presence of 10% dimethyl sulfoxide (Sigma-Aldrich®) were thawed and expanded in T75 flasks. For EVs collection, sub-confluent HLSCs were cultured in serum-free Minimum Essential Medium Eagle-Alpha Modification (α-MEM) (Euroclone) for 18 h, in HYPERFlask® (Corning®, VWR International, Milan, Italy). Cell debris and apoptotic bodies were removed by centrifugation at 3,000 g for 20 min and by microfiltration with 0.22 µm filters. Then, the supernatant was ultracentrifuged at 100,000 g for 2 h at 4°C (Beckman Coulter Optima™ L-90K, Brea, CA, USA), and the EV pellet was resuspended in serum-free culture medium, in the presence of 1% of dimethyl sulfoxide. HLSC-EVs size and concentration were determined using the NanoSight NS300 system (NanoSight Ltd., Amesbury, UK) supplied with a 405 nm laser, and the analysis was carried out by the Nanoparticle Tracking Analysis (NTA) 3.2 software [5]. EVs were stored at −80°C until use for successive experiments.
Cytofluorimetric analysis of EV was performed as previously described [5]. The analysis of 37 different exosomal surface proteins was conducted using a bead-based multiplex analysis kit (MACSPlex Exosome Kit, human, Miltenyi Biotec, Bergisch Gladbach, Germany), according to the manufacturer’s instructions [5, 21]. Briefly, EV samples were mixed with MACSPlex Exosome Capture Beads and kept overnight on an orbital shaker at room temperature, protected from light. Then, the resulting EVs-beads complexes were co-incubated with the anti-tetraspanins detection antibodies and rocked for 1 h, in the dark and at room temperature. After intensive washing with MACSPlex buffer to remove the non-specific binding, the CytoFLEX Flow Cytometer (Beckman Coulter) was employed to acquire at least 5,000 single-bead events per sample. Each capture bead subset population was detected and gated based on its respective fluorescence intensity. The measurement of the median fluorescence intensity (MFI) was performed using the CytExpert Software. The MFI value of a blank control (MACSPlex buffer + capture beads + detection antibodies) was subtracted from the MFI value of each capture bead subset, in order to remove background fluorescence intensity.
Transmission electron microscopy (TEM) analysis was carried out as previously described [5, 22]. EVs were spotted onto 200 mesh nickel formvar carbon-coated grids (Electron Microscopy Science, Hatfield, PA, USA). Once adhered to the grid, EVs were fixed using a solution of 2.5% glutaraldehyde and 2% sucrose, and then washed in distilled water. The negative staining was performed by incubating samples with Nano-W® and NanoVan® (Nanoprobes, Yaphank, NY, USA). Images were acquired using a Jeol JEM-1400 flash electron microscope (Jeol, Tokyo, Japan).
For western blotting analysis, protein lysates obtained from cells and EVs were processed as previously described [9]. Protein concentration was determined by bicinchoninic acid (BCA) Protein Assay Kit (Pierce™ Thermo Fisher Scientific, Waltham, MA, USA). Mini-PROTEAN® TGX™ Precast Protein Gels (4–20% gradient, Bio-Rad, Hercules, CA, USA) were loaded with protein samples (20 µg), under reducing conditions. Using the Trans-Blot® Turbo™ Transfer System (Bio-Rad), protein bands were transferred onto 0.2 µm nitrocellulose membranes, which were saturated in phosphate-buffered saline supplemented with 0.1% Tween-20 (PBS-T) and 5% non-fat milk. Then, membranes were probed at 4°C over-night with specific primary antibodies for exosomal markers, all used with a dilution of 1:1,000 in PBS-T supplemented with 5% bovine serum albumin: rabbit anti-CD81 (Cell Signaling Technology, Danvers, MA, USA), mouse anti-CD63 (Santa Cruz Biotechnology, Dallas, TX, USA), mouse anti-TSG101, and rabbit anti-CD9 (Abcam, Cambridge, UK). Primary antibody rabbit anti-GM130 (Abcam) was employed to exclude the presence of cell contaminants in EVs. After repeated washing, membranes were probed for 1 h at room temperature with anti-mouse or anti-rabbit peroxidase-conjugated secondary antibodies (Thermo Fisher Scientific), both diluted 1:5,000 in PBS-T. After incubation with the enhanced chemiluminescence substrate (SuperSignal™ West Femto Maximum Sensitivity Substrate, Thermo Fisher Scientific), the chemiluminescent signal was acquired using the ChemiDoc System (Bio-Rad).
In vivo murine model
In vivo experiments were carried out in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals, following the guidelines of Animal Research: Reporting of In Vivo Experiments (ARRIVE) and the mice were randomly assigned to the different experimental groups. All procedures were approved by the Italian Health Ministry (authorization number: 419/2016-PR). NASH was induced by continuously feeding the mice with a methionine and choline deficient (MCD) diet, as previously reported [4, 5, 23–26]. Ten weeks old severe combined immunodeficient (Charles River Laboratories, Wilmington, MA, USA) male mice were accustomed to a MCD diet (MP Biomedicals, Eschwege, Germany) by feeding a mixture of standard and MCD diet. After 1 week of mixed diet, the full MCD diet was administered to the mice. Starting at week 2 of the diet, 2.5 × 109 HLSC-EVs were intravenously injected twice a week (Figure 1A). Each mouse in the EV-treated group received a total of four EV injections (n = 8 mice), while mice in the NASH group were injected with vehicle alone (phosphate-buffered saline, n = 10 mice). Healthy mice (n = 8) were fed with the standard diet. All animals were sacrificed by cervical dislocation after deep sedation, at week 4 and liver tissue was recovered and maintained in RNA later at –20°C for subsequent RNA extraction and molecular analyses, as described below.
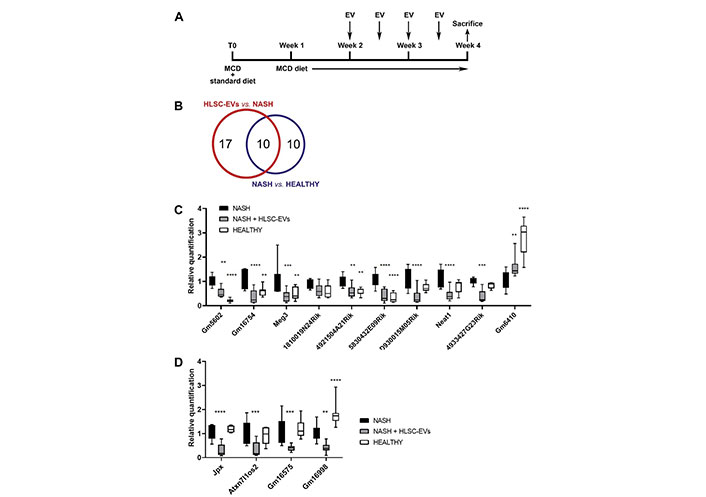
Inflammatory-related lncRNAs modulated in NASH mice that received HLSC-EVs or vehicles alone. A. Graphical representation of the in vivo experimental plan, showing the timings of MCD diet administration, of HLSC-EVs injection, and of sacrifice; B. Venn’s diagram representing the results of the screening with lncRNA array and showing the number of lncRNAs significantly modulated in each experimental group; C and D. real-time polymerase chain reaction (PCR) validation of selected lncRNAs on the RNA of NASH mice treated (n = 8) and not treated (n = 10) with HLSC-EVs. The RNA of healthy mice (n = 8) was used as control; C. lncRNAs whose expression was dysregulated by NASH and partially reverted towards baseline levels by HLSC-EV administration; D. lncRNAs not modulated by NASH but reduced by treatment with HLSC-EVs. LncRNA expression was normalized on the arithmetic mean of the housekeeping genes B2m and Rn7sk. NASH mice were considered reference control. Statistical analysis was performed using the two-way analysis of variance (ANOVA) test: ** P < 0.01; *** P < 0.001; **** P < 0.0001 vs. NASH mice. Neat1: nuclear enriched abundant transcript 1
In vitro model of HSC activation
The human hepatic immortalized stellate cell line (LX-2) [27] (Sigma-Aldrich®), was cultured in Dulbecco’s Modified Eagle’s Medium high glucose (4.5 g/L, Euroclone) in the presence of 2 nmol/L L-Glutamine (Lonza, Basel, Switzerland) and 2% fetal bovine serum, and used up to passage 6. After seeding at a density of 15,000 cells/cm2 in 6-well plates (Corning), LX-2 was maintained in serum-deprived medium over-night, with the addition of 0.2% of bovine serum albumin (Sigma-Aldrich®), to synchronize the cell culture. Incubation with transforming growth factor-beta 1 (TGF-β1, 10 ng/mL, purchased by Sigma-Aldrich®) for 6 h was performed to induce the activation of LX-2. Then, activated LX-2 was stimulated using a single administration of 50,000 (50k) EVs per cell, or a lower dose of 10,000 (10k) EVs per cell repeatedly administered once a day for 3 days, as described [9]. Cells were lysed after 24 h and 72 h of incubation with EVs to perform molecular analysis.
RNA extraction
For in vitro experiments, total RNA was collected from LX-2 using the commercially available miRNeasy mini kit (Qiagen, Valencia, CA, USA). For in vivo experiments, total RNA was obtained from liver tissue of healthy mice, NASH mice treated and not treated with HLSC-EVs using TRIzol™ reagent (Ambion, Thermo Fisher Scientific), and homogenized in a Bullet Blender instrument (Next Advance Inc., New York, NY, USA), as previously described [5]. The resulting RNA was diluted in RNase-free water and quantified using mySPEC spectrophotometer (VWR, Radnor, PA, USA).
LncRNA array
To perform the lncRNA screening, 1 µg of RNA isolated from the liver was converted into complementary DNA (cDNA) with RT2 First Strand Kit and lncRNA expression was analyzed using RT2 lncRNA PCR Array Mouse Inflammatory Response & Autoimmunity (LAMM-004ZC, Qiagen), following the manufacturer’s protocol. The QuantStudio™ 12K Flex Real-Time PCR instrument (Thermo Fisher Scientific) performed quantitative real-time polymerase chain reaction (qRT-PCR). The expression of 84 inflammation-related lncRNAs (Table S2) was compared among healthy mice (n = 3), NASH mice treated (n = 4) and not treated (n = 5) with HLSC-EVs, using GeneGlobe software (Qiagen). CT cut-off was set to 35 and the fold-regulation threshold was set to 2. The arithmetic mean of two housekeeping genes (B2m and Rn7sk) was used to normalize array data. Fold regulation expression with respect to the control group was calculated for all samples using the ΔΔCt method.
Real-time PCR analysis
To confirm the expression of specific lncRNAs, qRT-PCR was carried out on the RNA of healthy mice (n = 8), NASH mice treated (n = 8) and not treated (n = 10) with HLSC-EVs. For each sample, 2 µg of RNA were converted into cDNA and analyzed using specific primers (RT2 lncRNA PCR Assays provided by Qiagen and listed in Table S3) for murine lncRNAs selected from the array screening. Data analysis was performed using ExpressionSuite software (Applied Biosystems, Waltham, MA, USA). The arithmetic mean of two housekeeping genes (B2m and Rn7sk) was used to normalize qRT-PCR data.
To confirm the expression of lncRNA Meg3 in vitro, qRT-PCR was carried out as previously described [9]. Briefly, 5 ng of RNA from LX-2 were converted into cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems), were mixed with Power SYBR™ Green PCR Master Mix (Applied Biosystems) and specific oligonucleotide primers for human Meg3 (forward, 5’-TCCATGCTGAGCTGCTGCCAAG-3’; and reverse, 5’-AGTCGACAAAGACTGACACCC-3’) from previous publication [28] (100 nmol/L, obtained from MWG-Biotech, Eurofins Scientific, Brussels, Belgium). To normalize qRT-PCR data, the TATA binding protein (TBP) gene (primers: forward, 5’-TGTGCACAGGAGCCAAGAGT-3’; and reverse, 5’-ATTTTCTTGCTGCCAGTCTGG-3’) was used as housekeeping. For all samples, fold change expression with respect to control was measured using the ΔΔCt method.
Statistical analyses
Statistical analyses were conducted using the GraphPad Prism software v.8.0 and the results were displayed as mean ± standard deviation (SD). ANOVA with Dunnett’s multiple-comparisons test was applied for data analysis and P < 0.05 was considered significant.
Results
HLSC-EVs modulate inflammation-related lncRNAs in liver tissue of NASH mice
The therapeutic effect of HLSC-EVs on liver inflammation and fibrosis has been previously demonstrated in a murine model of NASH [5] (Figure 1A). Based on the positive results obtained in this MCD diet model, the expression of 84 inflammation-related lncRNAs was investigated in NASH mice subjected to treatment with EVs or with vehicle alone. The results of the lncRNA screening are summarized in Table S4 and Venn’s diagram is shown in Figure 1B.
By comparing the NASH group with the healthy group, 20 lncRNAs were found modulated in the NASH group (17 up-regulated and 3 down-regulated, as shown in Table 1). When the EV-treated group was compared with the NASH group, the expression of 27 lncRNAs (26 down-regulated and 1 up-regulated, as shown in Table 2) was found modulated by the EV treatment. Interestingly, among the lncRNAs differentially expressed in NASH mice, the expression of 10 lncRNAs (Tables 1 and 2) was partially restored to control levels by the treatment with EVs.
LncRNAs modulated in NASH mice in comparison with healthy mice. Fold regulation expression of each lncRNA with respect to healthy mice was calculated using the ΔΔCt method
| Symbol | Fold regulation |
|---|---|
| Gm5602 | 6.59 |
| 5830432E09Rik# | 5.75 |
| 4921504A21Rik | 4.07 |
| Meg3# | 3.43 |
| Gm16754# | 3.37 |
| A430108G06Rik | 3.08 |
| Gm14379 | 3.05 |
| 1810019N24Rik# | 2.58 |
| C230088H06Rik | 2.57 |
| 4933427G23Rik# | 2.53 |
| D930015M05Rik# | 2.35 |
| Neat1# | 2.3 |
| Gm15050# | 2.25 |
| Gm4117# | 2.15 |
| 9430037G07Rik | 2.14 |
| A930024N18Rik# | 2.13 |
| Chd3os | 2.03 |
| Dleu2 | –2.05 |
| 2310001H17Rik | –2.29 |
| Gm6410 | –2.98 |
# LncRNAs modulated both in NASH mice compared with healthy mice, and in HLSC-EV treated NASH mice compared with NASH mice
LncRNAs modulated in HLSC-EV treated mice in comparison with NASH mice. Fold regulation expression of each lncRNA with respect to NASH mice was calculated using ΔΔCt method
| Symbol | Fold regulation |
|---|---|
| Atxn7l1os2 | –4.16 |
| Jpx | –4.13 |
| 5830432E09Rik# | –3.48 |
| D930015M05Rik# | –3.29 |
| Gm16998 | –3.1 |
| Gm16575 | –3.07 |
| Gm16754# | –3.03 |
| Neat1# | –2.68 |
| Meg3# | –2.66 |
| 4933427G23Rik# | –2.54 |
| Dlx1as | –2.54 |
| Gm17354 | –2.43 |
| Gm15050# | –2.42 |
| Gm16892 | –2.41 |
| Gm17275 | –2.38 |
| Gm4211 | –2.35 |
| 5530601H04Rik | –2.3 |
| Gm4117# | –2.29 |
| 1810019N24Rik# | –2.24 |
| Firre | –2.2 |
| A330023F24Rik | –2.18 |
| Peg13 | –2.17 |
| Slc2a4rg-ps | –2.16 |
| A930024N18Rik# | –2.11 |
| Gm17586 | –2.04 |
| Gm17388 | –2.04 |
| Redrum | 2.33 |
# LncRNAs modulated both in NASH mice compared with healthy mice, and in HLSC-EV treated mice compared with NASH mice
It has been reported in the literature that some lncRNAs included in the screening are involved in chronic liver fibrosis [10]. In particular, the expression of Meg3 and Neat1 was up-regulated in NASH mice and significantly down-regulated by EV treatment. Instead, the expression levels of metastasis associated in lung adenocarcinoma transcript 1 (Malat1) and growth arrest-specific transcript 5 (Gas5) were slightly increased in NASH mice compared to healthy mice, but not affected by EV treatment (Table S4).
To confirm the results obtained from the lncRNA screening, the expression of the 14 lncRNAs reported in Table 3 was validated on larger samples. In particular, 7 lncRNAs dysregulated by NASH and partially restored by treatment with EVs (5830432E09Rik, Meg3, Gm16754, 1810019N24Rik, 4933427G23Rik, D930015M05Rik, Neat1), 3 lncRNAs dysregulated only in NASH group (Gm5602, 4921504A21Rik, Gm6410), and 4 lncRNAs modulated only by treatment with EVs (Atxn7l1os2, Jpx, Gm16998, Gm16575) were selected.
LncRNAs selected for real-time PCR validation
| Symbol | Dysregulated by NASH | Modulated by EV treatment |
|---|---|---|
| 5830432E09Rik | yes | yes |
| Meg3 | yes | yes |
| Gm16754 | yes | yes |
| 1810019N24Rik | yes | yes |
| 4933427G23Rik | yes | yes |
| D930015M05Rik | yes | yes |
| Neat1 | yes | yes |
| Gm5602 | yes | no |
| 4921504A21Rik | yes | no |
| Gm6410 | yes | no |
| Atxn7l1os2 | no | yes |
| Jpx | no | yes |
| Gm16998 | no | yes |
| Gm16575 | no | yes |
Among the lncRNAs selected by the array screening, the expression levels of 9 lncRNAs were up-regulated by NASH and down-regulated by EV treatment (Figure 1C), while the expression level of 1 lncRNA was down-regulated by NASH and up-regulated by EV administration (Figure 1C). In addition, the expression levels of 4 lncRNAs were not modulated by NASH but were strongly reduced by treatment with EVs (Figure 1D). These results confirmed what has been previously observed in the lncRNA screening on NASH mice treated and not treated with EVs.
HLSC-EVs down-regulate Meg3 expression in activated LX-2
Based on the results obtained by the lncRNA screening on NASH mice, subsequent investigation focused on Meg3, a lncRNA whose expression is dysregulated in NASH and restored to control levels by EV treatment. Meg3 expression was evaluated in an in vitro fibrosis model established using TGF-β1-activated LX-2 [9].
HLSC-EVs were characterized according to the International Society of EVs guidelines [29]. EVs expressed classical exosomal markers TSG101, CD9, CD63, and CD81, as shown by western blot analysis (Figure 2A) and cytofluorimetric analysis (Figure 2B). The mesenchymal origin of HLSC-EVs was confirmed by the expression of CD29, CD44, CD105, CD146, and the adhesion molecule melanoma-associated chondroitin sulfate proteoglycan (MCSP) (Figure 2B). Furthermore, the expression of epithelial markers EpCAM/CD326 and CD24, hematopoietic (CD45) and endothelial (CD31) markers, and human leukocyte antigen (HLA) class I and II was not detected on EVs (Figure 2B). NTA analysis showed that the average size of EVs was 173.3 nm ± 5.3 nm (Figure 2C) and TEM analysis confirmed that EV preparations contained intact nano-sized particles with double membrane (Figure 2D).
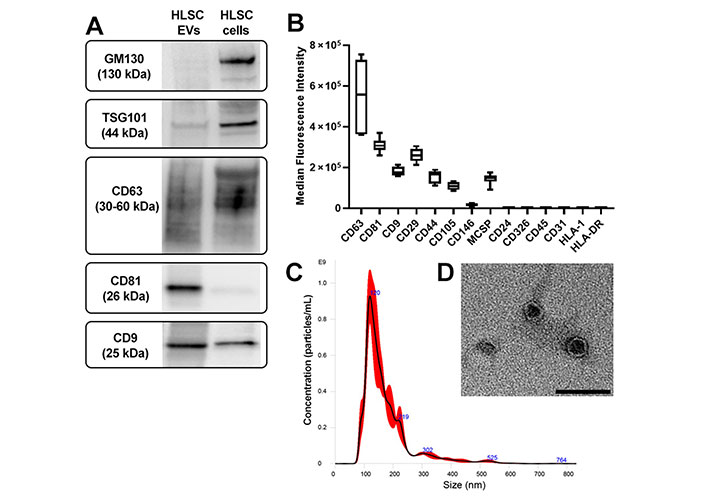
Characterization of HLSC-EVs. A. Western blot images of protein bands showing the expression of the typical exosomal markers TSG101 and tetraspanins (CD63, CD81, CD9) in EVs. As non-exosomal protein, the cis-Golgi marker GM130 is expressed only in cells (HLSCs); B. cytofluorimetric analysis of the phenotype of HLSC-EVs. Median allophycocyanin fluorescence values are reported for 14 selected bead populations after background. The graph shows the results of three different EV preparations; C. representative NTA graph showing EV concentration and size distribution; D. TEM image of EVs negatively stained with Nano-W® and NanoVan® (scale bar, 100 nm; magnification, 50,000×)
The setup for the in vitro experiments is shown in Figure 3A. The activation of LX-2 was induced by stimulation with TGF-β1. The effects of different doses and regimens of EV administration were tested on activated LX-2. The up-regulation of Meg3 expression levels was observed in TGF-β1-activated LX-2, compared to non-activated cells. Furthermore, the stimulation of activated LX-2 with EVs down-regulated the expression levels of Meg3 after 24 h (Figure 3B). Interestingly, the down-regulation of Meg3 is maintained after 72 h of incubation with the single EV administration (50k EVs/cell) (Figure 3C).
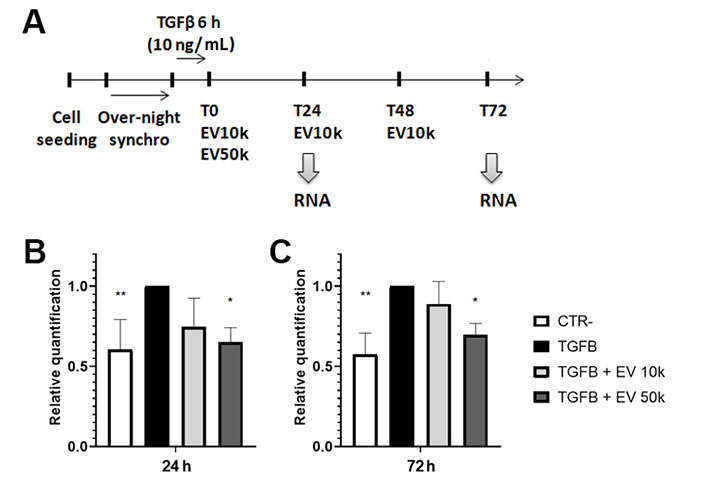
Modulation of lncRNA Meg3 by HLSC-EVs in HSCs. A. Graphical representation of the experimental plan to test in vitro the EV effects on activated HSCs. In the scheme are shown the timings of cell seeding, synchronization, activation with TGF-β1, EV treatment, and RNA purification from HSCs; B and C. histograms showing the expression level of Meg3 in HSCs after 24 h (B) and 72 h (C) of incubation with EVs, evaluated by real-time PCR. The housekeeping gene TBP was used to normalize gene expression levels. Activated HSCs and not treated with EVs were considered the reference control (TGFB). HSCs not activated with TGF-β1 were considered as negative control (CTR-). Statistical analysis was carried out on data collected from three independent runs, using the two-way ANOVA test: * P < 0.05; ** P < 0.01 vs. TGF-β1-activated cells
Discussion
HLSCs are a mesenchymal stromal cell-like population resident in the liver that was described for the first time in 2006 [30]. Along with a multipotent differentiating ability, HLSCs express mesenchymal and embryonic markers and retain remarkable therapeutic and pro-regenerative properties [30, 31]. In an in vivo model of NASH induced by MCD diet, a strong anti-inflammatory and anti-fibrotic effect of HLSC-EVs was observed at histological and molecular levels [5]. In particular, EV treatment down-regulated the expression levels of several fibrosis-related transcripts that were up-regulated by NASH. The present study reports that HLSC-EVs can also influence the expression levels of inflammation-related lncRNAs in the liver of NASH mice.
LncRNAs are involved in the regulation of proliferation, differentiation, development, apoptosis [32], and their aberrant expression is associated with the pathogenesis and progression of various human diseases [33], including liver fibrosis. Microarray analysis of the lncRNA expression profile in the liver of NAFLD patients identified more than 1,000 lncRNAs whose expression was dysregulated compared to normal liver [34]. For example, the liver expression levels of Malat1 correlated with NAFLD severity and were found up-regulated in patients with NASH [35, 36].
Besides liver biopsies, several lncRNAs associated with the development and progression of NAFLD have been also identified in in vivo mouse models, trying to investigate their complex mechanism of action [37–39]. In particular, in mouse models of NAFLD and NASH, the up-regulation of inflammation-related lncRNAs, such as Malat1 [40], H19 [41–44], HOX transcript antisense intergenic RNA (Hotair) [45], Gas5 [46, 47], and Neat1 [48–50], correlated with liver fibrosis, and their down-regulation inhibited the progression of NAFLD.
In the present study, the lncRNA expression profile in the liver of NASH mice treated and untreated with HLSC-EVs was explored by performing an array screening of 84 lncRNAs known to be associated with inflammation. The results of the lncRNA screening indicate that the expression of several inflammation-related lncRNAs is dysregulated in NASH livers. EV treatment can modulate the hepatic lncRNA expression profile, in particular by restoring to normal levels the expression of some lncRNAs dysregulated in NASH. Notably, real-time PCR validation of selected lncRNAs, carried out on larger samples, increased the number (from 7 to 10) of lncRNAs modulated by NASH and partially restored by treatment with EVs. To our knowledge, this is the first demonstration of the possibility of modifying the lncRNA expression profile of liver tissue after a chronic injury, using EVs derived from human mesenchymal stromal-like cells.
A dysregulation of the lncRNA expression profile has been also observed during HSC activation in vitro [51]. In addition, by comparing lncRNAs associated with the activated myofibroblastic state of LX-2 with differentially expressed lncRNAs in NAFLD patients, Gerhard and colleagues [52] have identified 91 lncRNAs shared between the two datasets. Based on this, one could speculate that some inflammation-related lncRNAs involved in the activation of HSCs may also contribute to the progression of hepatic fibrosis in NAFLD patients.
This study focuses in particular on the expression levels of Meg3, a lncRNA that has been observed up-regulated in NASH and restored to control levels by EV treatment. Here, for the first time Meg3 expression is investigated in an in vivo MCD diet-induced model of NASH and the reported results are in line with what has been observed in the liver of NAFLD and NASH patients [53, 54]. In NAFLD models in which mice are fed by a high-fat diet, Meg3 shows high expression levels in the liver that correlate with enhanced hepatic insulin resistance [55–57]. In particular, Meg3 induces an increase in forkhead box protein O1 expression [55], facilitates activating transcription factor 4 expression by acting as a competing endogenous RNA of microRNA (miR)-214 [56], and positively regulates early growth response proteins-2 by inhibiting miR-185-5p [57]. However, the role of Meg3 in NAFLD is debated, since other research groups indicate low levels of Meg3 in high-fat diet-induced NAFLD model [58–60]. In these studies, over-expression of Meg3 alleviates oxidative stress and lipid over-deposition, by positively regulating nuclear factor erythroid-2-related factor 2 [58], and by acting as competing endogenous RNA of miR-21 to regulate the expression of low-density lipoprotein receptor-related protein 6 [59]. Furthermore, over-expression of Meg3 in hepatocytes suppresses lipogenesis and inflammation by ubiquitinating enhancers of zeste homolog 2 and up-regulating sirtuin 6 [60]. As suggested by Cheng and colleagues [54], the induction of Meg3 in NAFLD and NASH patients might represent a compensatory mechanism. They demonstrated that Meg3 maintains glucose homeostasis and insulin signaling by protecting the hepatic endothelium against cellular senescence in obesity [54]. NAFLD patients frequently show insulin resistance, which may promote the reactivation of Meg3 to maintain glucose homeostasis. More in-depth studies are definitely needed to elucidate the regulation and mechanisms of action of Meg3 in the development and progression of fibrosis in NASH. However, based on evidence reported in this study, together with the controversial data reported in the literature, it is possible to speculate that Meg3 could display different mechanisms of action depending on the in vivo model considered and on the progression of liver fibrosis.
Recently, single-cell transcriptomic analysis in fibrotic livers of mice treated with carbon tetrachloride identified 8 different subpopulations of HSCs, each with a unique transcriptomic profile [61]. Interestingly, Meg3 was found among the top expressed genes of the largest HSC subpopulation with the typical phenotype of mature myofibroblasts, suggesting an important function of this lncRNA in activated HSCs. The evaluation of Meg3 expression in in vitro activated HSCs, showed a similar trend to that obtained in the in vivo model of NASH, induced by MCD diet. In fact, an increase of Meg3 expression levels was detected in TGF-β1-activated LX-2 compared to quiescent cells, while treatment with EVs partially restored its expression to baseline levels. However, previous studies have shown a down-regulation of Meg3 in LX-2 in response to TGF-β1, and an inhibitory effect of Meg3 enforced expression on LX-2 activation [62, 63]. These differences could be attributed to different cell culture conditions, which do not permit to maintain the quiescent state of HSCs for a long time. For instance, cultivation in low serum conditions (2%) should be preferable to preserve the quiescent state of LX-2, because higher serum conditions (10%) have been shown to induce the activated phenotype in LX-2 [64]. Besides, lacking the surrounding hepatic microenvironment, two-dimensional culture systems do not accurately reproduce the pathophysiological mechanisms of fibrogenesis and inflammation occurring in NASH [65]. For these reasons, three-dimensional culture systems of different populations of liver cells (e.g., hepatocytes, HSCs, and Kupffer cells) could be a preferable tool to study in vitro the expression profile and the mechanism of action of lncRNAs during HSC activation. Moreover, silencing of Meg3 in activated LX-2, together with the identification of its targets in LX-2, might be possible strategies to better define the role of this lncRNA in the reversal of LX-2 activated phenotype.
In conclusion, HLSC-EVs administration in NASH mice induced an attenuation of hepatic fibrosis, by regulating several genes [5] and lncRNAs associated with inflammation and fibrosis. Here it has been demonstrated that EVs derived from a human stem cell population might modify the lncRNA expression profile of the hepatic injured tissue. In particular, EV treatment can restore baseline levels of those inflammation-related lncRNAs dysregulated in NASH and activated LX-2, indicating their possible involvement in the beneficial effect of HLSC-EVs in liver fibrosis. However, since NASH mice were treated after a short period of exposure to the toxic insult, it is not possible to conclude that HLSC-EVs administration may also be effective in reversing the process of NASH-induced liver cirrhosis.
A more in-depth investigation of the molecular mechanisms underlying the regulation of the lncRNAs modulated by EVs could help to develop new strategies to hinder the progression of hepatic fibrosis in NASH.
Abbreviations
| ANOVA: | analysis of variance |
| cDNA: | complementary DNA |
| EVs: | extracellular vesicles |
| HLA: | human leukocyte antigen |
| HLSCs: | human liver stem cells |
| HSCs: | hepatic stellate cells |
| lncRNA: | long non-coding RNA |
| LX-2: | human hepatic immortalized stellate cell line |
| Malat1: | metastasis associated in lung adenocarcinoma transcript 1 |
| MCD: | methionine and choline deficient |
| Meg3: | maternally expressed gene 3 |
| MFI: | median fluorescence intensity |
| miR: | microRNA |
| NAFLD: | non-alcoholic fatty liver disease |
| NASH: | non-alcoholic steatohepatitis |
| Neat1: | nuclear enriched abundant transcript 1 |
| NTA: | Nanoparticle Tracking Analysis |
| PBS-T: | phosphate-buffered saline supplemented with 0.1% Tween-20 |
| PCR: | polymerase chain reaction |
| qRT-PCR: | quantitative real-time polymerase chain reaction |
| TEM: | transmission electron microscopy |
| TGF-β1: | transforming growth factor-beta 1 |
Supplementary materials
The supplementary material for this article is available at: https://www.explorationpub.com/uploads/Article/file/100525_sup_1.pdf.
Declarations
Author contributions
G Chiabotto: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing—original draft. EC: Formal analysis, Investigation, Methodology, Validation, Visualization. CP: Investigation. MBHS: Investigation, Methodology, Formal Analysis. MC: Methodology. G Camussi: Funding acquisition, Resources, Writing—review & editing. SB: Conceptualization, Formal analysis, Funding acquisition, Project administration, Resources, Supervision, Visualization, Writing—review & editing. All authors have read and agreed to the published version of the manuscript.
Conflicts of interest
G Camussi is a component of the scientific advisory board of Unicyte and is named inventor in a related patent (WO2006126219-A1). The other authors declare that they have no conflict of interest.
Ethical approval
HLSCs were generated from one donor liver. The permission was obtained by the National Transplant Center in accordance with Italian legislation. The animal study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Italian Health Ministry (authorization number: 419/2016-PR, 26/04/2016).
Consent to participate
Not applicable.
Consent to publication
Not applicable.
Availability of data and materials
All datasets generated for this study are included in the manuscript and the supplementary files.
Funding
This research was funded by grant “Terapie avanzate per processi fibrotici cronici” [Progetto EVER, codice domanda 320-39], by grant RiLo-2020, and by Unicyte AG (Oberdorf, NW, Switzerland). The funders had no role in the study design, data collection, analysis and interpretation of the data, the decision to publish, or the preparation of the manuscript.
Copyright
© The Author(s) 2023.