Affiliation:
1Department of Radiation Oncology and Molecular Radiation Sciences, Image-Guided Therapy Lab, Johns Hopkins University, Baltimore, MD 21218, USA
Email: kai@jhu.edu
ORCID: https://orcid.org/0000-0002-8121-0258
Affiliation:
2Department of Social Medicine, University of North Carolina-Chapel Hill, Chapel Hill, NC 27599, USA
ORCID: https://orcid.org/0009-0009-9303-7382
Affiliation:
1Department of Radiation Oncology and Molecular Radiation Sciences, Image-Guided Therapy Lab, Johns Hopkins University, Baltimore, MD 21218, USA
Affiliation:
1Department of Radiation Oncology and Molecular Radiation Sciences, Image-Guided Therapy Lab, Johns Hopkins University, Baltimore, MD 21218, USA
Affiliation:
1Department of Radiation Oncology and Molecular Radiation Sciences, Image-Guided Therapy Lab, Johns Hopkins University, Baltimore, MD 21218, USA
Explor Digit Health Technol. 2024;2:302–312 DOI: https://doi.org/10.37349/edht.2024.00030
Received: April 30, 2024 Accepted: September 25, 2024 Published: November 12, 2024
Academic Editor: Mohammad Reza Saeb, Gdansk University of Technology, Poland
The article belongs to the special issue Cancer Diagnosis in the Digital Age
Aim: In lung cancer research, AI has been trained to read chest radiographs, which has led to improved health outcomes. However, the use of AI in healthcare settings is not without its own set of drawbacks, with bias being primary among them. This study seeks to investigate AI bias in diagnosing and treating lung cancer patients. The research objectives of this study are threefold: 1) To determine which features of patient datasets are most susceptible to AI bias; 2) to then measure the extent of such bias; and 3) from the findings generated, offer recommendations for overcoming the pitfalls of AI in lung cancer therapy for the delivery of more accurate and equitable healthcare.
Methods: We created a synthetic database consisting of 50 lung cancer patients using a large language model (LLM). We then used a logistic regression model to detect bias in AI-informed treatment plans.
Results: The empirical results from our synthetic patient data illustrate AI bias along the lines of (1) patient demographics (specifically, age) and (2) disease classification/histology. As it concerns patient age, the model exhibited an accuracy rate of 82.7% for patients < 60 years compared to 85.7% for patients ≥ 60 years. Regarding disease type, the model was less adept in identifying treatment categories for adenocarcinoma (accuracy rate: 83.7%) than it was in predicting treatment categories for squamous cell carcinoma (accuracy rate: 92.3%).
Conclusions: We address the implications of such results in terms of how they may exacerbate existing health disparities for certain patient populations. We conclude by outlining several strategies for addressing AI bias, including generating a more robust training dataset, developing software tools to detect bias, making the model’s code open access and soliciting user feedback, inviting oversight from an ethics review board, and augmenting patient datasets by synthesizing the underrepresented data.
According to the American Cancer Society [1], lung cancer is the second most common type of cancer in the United States. In 2024, an estimated 116,310 men and 118,270 women will be diagnosed with lung cancer, meaning that the disease affects both genders relatively equally [1]. Not only is lung cancer one of the most common cancers in both the U.S. and global context, but it is also the deadliest, with the highest mortality rates of any other cancer worldwide [2]. However, mortality rates and treatment plans vary depending on the type of lung cancer and the stage at which it is detected.
In medical nosology, lung cancer is refined into two types: small cell lung cancer (SCLC) and non-SCLC (NSCLC). SCLC is a faster-growing type of lung cancer, but tends to be less common, representing fewer than 1 out of 10 new lung cancer cases [3]. NSCLC, on the other hand, is slower growing but is more common, accounting for approximately 9 out of 10 lung cancer diagnoses. In terms of classification-specific treatment plans, for SCLC, chemotherapy is the gold standard, whereas for NSCLC, assuming that the cancer has not become metastatic, surgery is often the first course of action [4]. The modality of treatment is also determined by the stage of advancement, with early-stage detection and localization of the disease dictating one protocol and late-stage or metastatic lung cancer calling for another. Despite concerning mortality and morbidity rates, however, AI shows promise in augmenting both the detection and treatment plans of lung cancer of all types.
AI in its various forms is becoming increasingly more useful for a variety of healthcare purposes, from performing fetal ultrasounds [5] to training medical students and residents [6], to reviewing patient records [7]. It is therefore likely not surprising then that AI has come to play a central role in lung cancer research [8, 9].
As it concerns lung cancer, specifically, AI has proven beneficial in several regards. For one, AI can help enhance the accurate detection of lung cancer by improving image quality for precise tumor targeting, thus facilitating early detection and drastically improving treatment outcomes. To illustrate, Ardila et al. [10] found that when using AI to detect lung cancer in a database of over 40,000 computed tomography (CT) scans, the system’s ability to analyze entire 3D scans as opposed to just a series of 2D slices, rendered it more accurate than a human reader. This 3D perspective furthermore allowed the AI system to look for tumors in unexpected areas, further contributing to the diagnostic advantages it affords. In fact, the relative advantages AI holds over a radiologist have even led some to conclude that AI effectively “Beats Radiologists for Accuracy in Lung Cancer Screening” [11].
Additionally, AI supports dynamic treatment response assessment and outcome predictions, enabling more personalized and effective disease management strategies for lung cancer patients. AI has the potential to save more lives, not just with respect to more accurate detection rates, but with the ability to reach greater numbers of patients. From a pragmatic standpoint, AI may serve as an instrumental supplement to the radiology and oncology workforces. The American College of Radiology reports an ongoing global radiology labor shortage [12]. Such shortages are moreover exacerbated during times of health crises, as was witnessed with the COVID-19 pandemic. As an assistive technology, however, AI is poised to function as an auxiliary to clinicians in these fields, effectively ensuring that labor shortages do not prevent patients from receiving potentially life-saving care.
Despite the manifold advantages accompanying the use of AI in clinical settings, though, it is not without its own set of drawbacks, with bias being foremost among them. Bias in the use of AI within health settings has become an object of debate among both clinicians and researchers alike. To join this ongoing debate, and to provide further guidance as to how AI may be accurately and equitably applied in healthcare settings, we created a synthetic database consisting of 50 lung cancer patients using a large language model (LLM). We then used a logistic regression model to detect bias in AI-informed treatment plans. Our experiment was guided by the following two research questions:
To what extent does AI bias impact the accuracy of defining appropriate treatment categories for patients with various demographic backgrounds and types of lung cancer?
In what regard(s) is bias present? In other words, which features of the patient case were more susceptible to bias?
The article begins with our situating the present study within the larger scholarly discourse on the numerous benefits AI can bring to healthcare delivery, as well as how these benefits are often mitigated by different forms of bias. In the following section, we account for our methodological approach and describe our experiment design, before then presenting our results. The next section then proceeds to interpret said results. We conclude by reflecting on the implications of these results in terms of larger discussions of health equity and then examine how these pitfalls of AI may be overcome.
Before delving into the applications of AI in healthcare, however, an operating definition must first be established. “AI” is a broad term encompassing a variety of processes and applications, each with a different end goal. “AI” refers to tasks that a computer can perform with little to no assistance from human agents. Subsumed within the more inclusive category of AI is a subcategory of technologies that involve machine learning (ML). Simply put, “ML” denotes computer systems that learn from experience. Deep learning (DL) is one specific type of ML technology and the one that will be analyzed in this article. Coccia [13] defines DL as “a family of computational methods that allow an algorithm to program itself by learning from a large set of examples that demonstrate the desired behavior, removing the need to specify rules explicitly”. DL simulates how the human brain processes information through the creation of synthetic neural networks.
Regardless of the AI type, though, AI technologies as a whole have witnessed a preponderance of growth in the healthcare domain, particularly in the diagnosis and treatment of lung cancer. Coccia [14] claims that the growth of AI has occurred alongside and in tandem with growth in quantum technologies, the latter of which have played an increasingly important role in lung cancer detection and treatment via medical imaging. Given the symbiotic and synergistic relationship between AI and quantum technologies, investigation into one area inferably stands to benefit the other, in what the author conceptualizes as a cross-fertilization of innovation. He notes that there is an area of significant convergence between AI technologies and quantum technologies, and this study seeks to insert itself at this point of convergence.
Despite its growth, however, a primary reason that AI has not been more thoroughly integrated into clinical workflows for lung cancer treatment is that its accuracy remains inconclusive. The accuracy of AI in detecting thoracic cancer and assigning appropriate treatment categories based on those findings varies widely and lacks sufficient evidence to make determinations regarding its efficacy. That is why some, like Yang et al. [15], have issued a call for researchers to develop a more systematic analysis, and this study seeks to respond to this call by attempting to gauge the accuracy of AI for lung cancer detection and treatment.
Other barriers to improving the use of AI in lung cancer treatment lie with difficulty confirming its findings. Looking specifically at the use of AI for lung nodule detection via CT, Sourlos et al. [16] note that this technology often serves as an auxiliary one, not to be used in lieu of a qualified radiologist. For example, the authors conclude that how well AI will perform when it comes to detecting lung nodules in CT scans will depend, in large part, on whether it is enlisted as the first, second, or third reader of these images. Furthermore, evaluating the accuracy of AI in its ability to effectively detect lung cancer nodules is also contingent upon biopsy results for confirmation. These biopsies, considered the gold standard for confirmation of disease, are difficult to obtain, making evaluating the accuracy of AI in such applications equally difficult.
But accuracy and bias are two sides of the same coin. In other words, an inaccurate model is often a biased one. Tasci et al. [17] define the problem with AI bias as this: “If the bias is high, the model cannot capture the pattern (e.g., essential features) of the data, it misses the relations between the features and targets, and the learning model cannot generate appropriate predictions on testing data”. As one could therefore infer from this statement, AI bias largely stems from the quality and robustness of the data it is trained on, whether that is electronic medical health records (EMHR), insurance claims data, or medical device readings [18].
AI bias may assume many forms. In their review of bias and class imbalance in oncologic data, Tasci et al. [17] point out such AI bias may be broken down into two categories. The first of these categories of bias is clinical in nature and pertains to the over- or under-representation of certain patient populations. It is in this regard, and within the context of cancer, specifically, that the use of AI can exacerbate existing health disparities. To illustrate, consider Guo et al.’s systematic review [19] of the literature on skin cancer. The authors found that of the 136 studies included in their dataset, the race/ethnicity of the subject (39,820) was disclosed only 8.82% of the time. Therefore, if an AI model was to be trained on such an incomplete dataset, it could overlook this patient population with respect to cancer detection, thereby worsening health outcomes for them [20].
In addition to AI bias along the lines of race and ethnicity, it may also demonstrate bias according to other patient features, such as gender. In their study of AI-assisted image-based diagnoses for 12 different thoracic diseases, Larrazabal et al. [21] examined how an imbalanced dataset could lead to skewed diagnostic results. The authors used X-ray images, a well-regarded classifier, and a training dataset with a gender balance of 25%/75%, women to men. Not surprisingly, the model performance was significantly lower than that of one trained on a more balanced dataset.
Tasci et al. [17] also note that AI bias in the analysis of oncologic data may also be present in the heterogeneity of the disease. In other words, outside of patient demographics, the pathological classification of the disease can also be a significant source of AI bias, with AI being more adept at diagnosing some diseases than others. If, as these studies show, diagnostic AI is only as good as the dataset it is trained on, then it would be less effective in accurately detecting less common disease types and subtypes, as these would be less represented in the dataset. This line of reasoning holds important implications for lung cancer therapy because, as previously described, certain types of lung cancer (e.g., NSCLC and SCLC) are more or less common than others. If these types are underrepresented in the AI training set, this could lead to biases in diagnosis and therapy.
As Figure 1 below illustrates, treatment plans are developed in accordance with disease type and stage of progression. In the case that this figure represents, the disease type was classified as NSCLC, for which radiotherapy is a common course of action. Should an AI model fail to accurately classify the disease type, the prescribed treatment plan could be ineffective at best, and at worst, outright harmful.
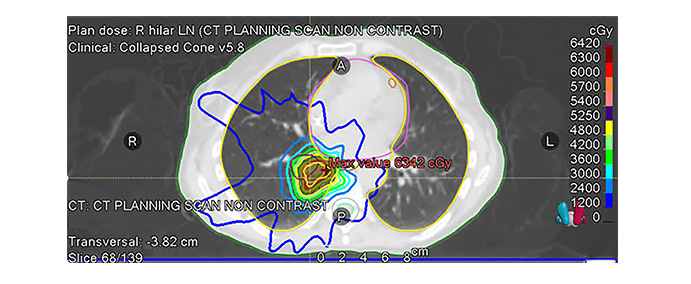
An example radiotherapy plan of a real lung cancer patient. The radiation isodose line is optimized to achieve tumor coverage and normal tissue sparing. Over 60% of patients with non-small cell lung cancer (NSCLC) require radiotherapy. Collected data was authorized through Johns Hopkins University’s Institutional Review Board’s guidelines for retrospective patient data use. Ethics committee name: JHU IRB; approval code: IRB-1 (retrospective patient data); approval date: 3/18/2020
In sum, while AI in healthcare—lung cancer diagnosis and treatment in particular—has consistently demonstrated improved health outcomes for some patient populations, it nonetheless represents the proverbial double-edged sword. One of the most significant drawbacks to the effective application of AI for lung cancer lies with the bias inherent in it. As this review has shown, the origin of such bias is not singular in nature; bias may occur along the lines of race or ethnicity, gender, or even disease type if the dataset the AI model is trained on is imbalanced. To systematically and empirically investigate the conditions that give rise to AI bias and the various ways that it may manifest, we developed our own experiment based on a synthetic patient database. The details of the data for analysis, as well as our methodological approach, are presented in the section that follows.
To analyze AI bias in recommendations for lung cancer treatment protocols, we created a synthetic database consisting of 50 patients using an LLM (Claude 3 Opus). A sample medical report items list was developed and selected as the prompt for the LLM. The report items included synthetic patient name, date of birth (DOB), stage of progression, treatment prescribed, radiology report with scan dates, tumor size, and location, pathology report with lymph node involvement, pathologic staging, treatment course, and follow-up visit report. While Figure 1 above was derived from real patient data, the data analyzed in this article was generated manually, meaning that no real data was used. Figure 2 below shows the sample distribution of this manufactured data, and is followed by an in-depth breakdown of the specific characteristics of this database.
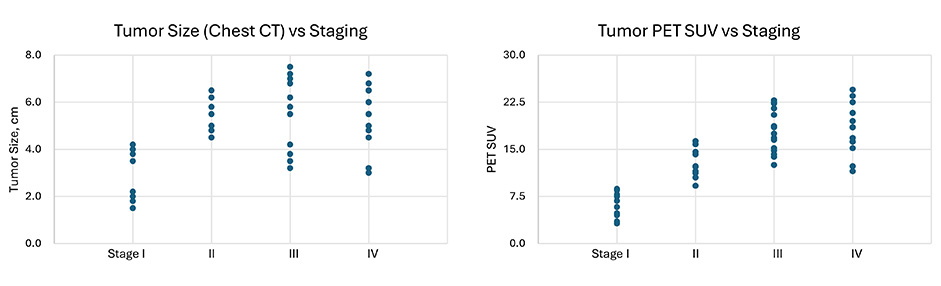
Distribution of 50 LLM-generated sample cases in terms of tumor size and PET SUV values vs different stages. LLM: large language model; PET: positron emission tomography; SUV: standard uptake value
Our selection of the specific features for our database was also informed by current practices using biomarkers to deliver immunotherapy for lung cancer. Recent advancements in lung cancer treatment have increasingly focused on immunotherapy, particularly through the exploitation of biomarkers such as programmed death ligand 1 (PD-L1) [22], which has been pivotal in managing NSCLC. PD-L1 is currently the only immunotherapy biomarker with an approved treatment for NSCLC; it enhances patient selection for therapies that block this pathway, thereby inhibiting tumor growth and progression. Immune checkpoint inhibitors (ICIs) directed against programmed cell death 1 (PD-1) and PD-L1 have markedly improved survival for patients with advanced NSCLC. Additionally, the utilization of other critical genomic biomarkers such as EGFR, ALK, and NTRK has been standardized, offering targeted therapies that significantly improve outcomes by personalizing treatment approaches based on individual genetic profiles. Therefore, in our database, we have included PD-L1 expression as a feature and variable of analysis as well.
The database comprises detailed records for 50 lung cancer patients, systematically organized across multiple dimensions to facilitate comprehensive analyses. Demographic variables include age, captured as a continuous variable, and gender. Clinical data features tumor size, measured in centimeters and recorded from chest CT scans, and PET (positron emission tomography) SUV (standard uptake value) values, which quantify the metabolic activity of the tumor. Cancer staging is thoroughly documented and is presented in two formats: a detailed traditional stage (I–IV) and a simplified categorization (stages 1–4), which simplifies the dataset for trend analysis and easier statistical handling. Treatment data encapsulates the type (surgery, chemotherapy, radiation, immunotherapy) and specifics, including drug names and radiation doses, the latter quantified in Gray (Gy). Biomarkers are extensively covered, with PD-L1 expression quantified as tumor proportion score (TPS) percentages. Performance status is rated using the Eastern Cooperative Oncology Group (ECOG) scale, ranging from 0 (fully active) to 5 (deceased). Complication severity following treatment is cataloged on a scale from mild to severe, and time to progression or recurrence is measured in months, providing a timeline of patient response post-treatment. Follow-up data includes the duration of follow-up in months and survival status, allowing for longitudinal outcome analysis. The specific features of this synthetic database are as follows:
Age distribution: The median age of patients was 58 years, with a range from 44 to 68 years.
Tumor size distribution: The average tumor size was 5.0 cm, with a range from 1.5 cm to 7.5 cm.
Tumor type distribution: 74% of the synthetic cases were diagnosed as adenocarcinoma and 26% were diagnosed with squamous cell carcinoma.
PET SUV distribution: The average PET SUV was 14.6, with a range from 3.2 to 24.5. PD-L1 expression: Among patients with available PD-L1 data, 36% had high PD-L1 expression (TPS ≥ 50%), while 64% had unknown.
In addition to classifying the dataset according to patient demographic information, tumor size and type, and stage of progression, we also differentiated the data according to treatment categories. The initial unique treatment categories constructed were as follows: lobectomy, systemic therapy with pembrolizumab, lobectomy followed by adjuvant chemotherapy, concurrent chemoradiation followed by immunotherapy, systemic therapy with carboplatin, pemetrexed, and pembrolizumab, neoadjuvant chemotherapy followed by lobectomy and adjuvant radiation systemic therapy with carboplatin, pemetrexed, and bevacizumab, stereotactic body radiation therapy (SBRT), systemic therapy with carboplatin, paclitaxel, and bevacizumab, wedge resection, and segmentectomy. After consolidation of the categories, we recategorized the cases into the classes of surgery, immunotherapy, radiation therapy, chemotherapy, or a combination of these.
Once we had established the above-described clinical feature variables, we then employed a logistic regression model to predict treatment categories based on this information. The logistic regression model was implemented using the Scikit-learn library in Python. For model inputs, key features included the clinical stage of cancer, tumor size measured via chest CT, and PET SUV values. The reason we selected these features is because previous studies, such as by Tsutani et al. [22], for instance, demonstrate the importance of these variables. These features were standardized using Scikit-learn’s StandardScaler to normalize the data. The dataset was divided into a training set (80%) and a testing set (20%). The logistic regression model was evaluated using accuracy as the primary metric. Accuracy is defined as the proportion of correct predictions over the total number of predictions made.
In terms of the overall model performance, the trained logistic regression model achieved an accuracy rate of 90%, a precision rate of 68.8%, a recall rate of 75%, and an F1 score of 0.714 on the testing set. The accuracy is calculated as (True Positive + True Negatives)/Total Incidences. The precision is calculated as True Positive/(True Positive + False Positives). The recall is calculated as True Positives/(True Positives + False Negatives). The F1 score is calculated as 2 × (Precision × Recall)/(Precision + Recall).
It is noteworthy, however, that the model’s performance varied across two different patient subgroups, the first of which is age. The model exhibited lower accuracy for patients younger than 60 years (82.7%) compared to those 60 years and older (85.7%) in the full datasets. We repeated the analysis by reducing the data from 50 to 20 from the synthetic database. When analyzed in the reduced datasets, the model was less accurate for patients younger than 60 years (75.0%) compared to those 60 years and older (87.5%). The results of these analyses are presented in Table 1 below.
Results of the large language model (LLM) on the synthetic patient dataset
| Category | Subcategory | Accuracy rate |
|---|---|---|
| Patient age | < 60 years | 82.7% |
| ≥ 60 years | 85.7% | |
| Disease type | Adenocarcinoma | 83.7% |
| Squamous cell carcinoma | 92.3% |
As Table 1 shows, discrepancies also occurred according to histological subtype with the full and reduced datasets. Our results indicate that the model was less adept in identifying treatment categories for adenocarcinoma, with an accuracy rate of 83.7%. When it came to predicting treatment categories for squamous cell carcinoma, however, the model performed better, with an accuracy rate of 92.3%. We present a summary of these results in the form of a strengths, weaknesses, opportunities, and threats (SWOT) analysis, summarized in Table 2 below.
SWOT analysis for AI in lung cancer treatment protocol recommendation
| Strengths | Weaknesses | Opportunities | Threats |
|---|---|---|---|
| Precision and accuracy: AI algorithms can analyze complex medical imaging and genetic data to identify subtle patterns for more precise and accurate treatment recommendations. | Algorithm bias: AI models can inherit biases from training data, which may lead to skewed recommendations if the data is not representative of diverse patient populations. | Continuous advancements in personalized medicine: AI has the potential to advance personalized medicine in lung cancer by analyzing individual patient data to tailor treatments. | Ethical and legal concerns: The ethical issues that accompany certain decisions and the potential legal implications of AI errors, pose significant challenges. Human physicians will need to review and sign off on patient charts in the current clinical setting. |
| Speed of diagnosis: AI can rapidly process and analyze large datasets, significantly reducing time expenditures. | High implementation costs: Developing, testing, and implementing AI systems for lung cancer treatment can be costly, requiring significant investment in technology and clinician expertise. | Integration with emerging technologies: Combining AI with emerging technologies like genomics can lead to a better understanding of lung cancer at the molecular level. | Technological disparities: There may be disparities in access to AI technologies between high and low-resource settings, potentially widening health inequities. |
| Consistency: AI systems provide consistent recommendations based on learned data, reducing variability in treatment suggestions among different oncologists. | Dependency on data quality: The effectiveness of AI recommendations is highly dependent on the quality and comprehensiveness of the data used, including historical treatment outcomes and patient demographics. | Global reach: AI can extend expert-level lung cancer treatment recommendations to underserved regions, improving outcomes where the access to oncologists is limited. | Resistance from healthcare professionals: There may be resistance to AI recommendations from healthcare professionals who are skeptical of replacing traditional clinical judgment with algorithmic decisions. |
In their study of AI applications for lung digital pathology, Viswanathan et al. [23] note that AI errors in such applications tend to originate from the training phase; the results of our study are consistent with such a claim. As expected, the model’s performance shows less dependency on the age categories (younger than 60 years vs 60 years and older) when there are larger training datasets when we use the full database (50 cases) compared to the reduced database (20 cases). Most real-world lung cancer diagnoses occur after age 65, with the median age of diagnosis being 66 [24]. The median age for our dataset, however, was 58, a fact that thus highlights the importance of a sound training set. This becomes problematic when one takes into account that age is a primary social determinant of health (SDOH) [25]. If a training dataset is not well-aligned with epidemiological realities and patient needs, then it could exacerbate existing health disparities among the aging population.
It is also significant that the model was more accurate in predicting treatment recommendations for squamous cell carcinoma, compared with adenocarcinoma, an NSCLC, and arguably the most common variety of lung cancer [26]. In this case, the training data set did not reflect the incidence rate of this disease in the general population. Such a finding is concerning, as adenocarcinoma is the most prevalent NSCLC type, comprising an estimated 30% of cases [26]. This means that if this model was enlisted in actual clinical settings, it would presumably fail to identify a significant portion of the patient population.
While this study yielded noteworthy findings, it is not without its limitations. The most pronounced limitation is that it is based on a small dataset. With a dataset consisting of only 50 patients, the findings extrapolated should not be regarded as decisive. It is also worth pointing out that this dataset drew from an exclusively Western (i.e., American) patient sample. Yet as Carini et al. [27] advise, inclusive algorithms that utilize data from global populations exhibit greater accuracy when it comes to predicting cancer across diverse populations. In light of these limitations, then, future studies wishing to build upon the present one may want to broaden the dataset to not only include a greater number of cases, but also expand the geographical scope from which patient data was sampled to likewise broaden the scope of the generalizability of the findings. Doing this would work to fulfill the call presently issued in research on AI in healthcare applications to develop ML models in healthcare “so that protected and non-protected demographic groups derive equal clinical benefits performing equally between the groups”.
Additionally, though the current study focused on logistic regression due to its suitability to demonstrate the AI bias inherent in small datasets and their interpretation, several other ML models could potentially be applied to investigate this problem in future research. Other potential models can be used but are out of the scope of this study; these include support vector machines (SVM) if the data exhibits a high-dimensional space more effectively separable through hyperplanes or neural networks if large-scale data becomes available or imaging data is integrated (e.g., radiomics).
Another limitation of this study, which likewise presents as an opportunity for future work to consider, is that we only analyze AI’s ability to perform two tasks: data mining and data analysis. Therefore, the results of this study cannot be used to make claims regarding the efficacy or accuracy of AI for other applications. Considering that human interpretation of images is subject to several limitations, He et al. [28] have called for the introduction of AI to analyze raw data from medical images in a “signal-to-knowledge” approach that bypasses the image reconstruction stage in a traditional “signal-image-knowledge” model. The promise of AI vis-à-vis medical imaging will ideally prompt future work to examine AI bias in the context of computer vision. This would hold important implications for lung cancer therapy in particular, as traditional diagnostic and therapeutic technologies such as detecting molecular biomarkers of lung biopsies and blood testing are accompanied by several disadvantages (e.g., invasive surgical interventions, prolonged wait times for test results) [29]. AI for computer vision and interpretation of medical imaging is poised to circumvent these limitations, and so represents a worthy area of further study.
In acknowledging these drawbacks to the use of AI in lung cancer diagnosis and treatment, a few relevant points of consideration for future AI applications emerge. The first reminds researchers and clinicians to engage in diligent reporting in patient records so as to help create a more robust natural data set for AI to learn from. With greater information supplied by actual clinical practices and observations, the diagnostic and therapeutic findings derived from AI can more realistically mirror clinical experiences. The second key takeaway from this brief experiment is that AI does not only demonstrate bias towards specific patient populations, but towards disease type, as well. Training the model on disease types with the highest incidence rates will help ensure that there are fewer instances of a failure to diagnose, and will ensure that greater numbers of individuals receive the care they need.
To address these relative shortcomings, we recommend implementing several of the following practices. The first practice involves developing software tools to detect and monitor AI bias. The second would seek out user feedback and invite an ethics review board to perform a manual audit. Relatedly, making the model and code open access would allow for such feedback, thus evolving the model through continuous improvement and imbuing the research process with transparency. Finally, we would recommend augmenting patient datasets by synthesizing the underrepresented data to improve the model’s performance. We believe that by taking such steps, AI can lead to innovation, not just in lung cancer detection and treatment, but for cancer care in general. These innovations may, in turn, directly translate to better patient health outcomes, improved quality of life, and ultimately, lives saved.
CT: computed tomography
DL: deep learning
LLM: large language model
ML: machine learning
NSCLC: non-small cell lung cancer
PD-L1: programmed death ligand 1
PET: positron emission tomography
SCLC: small cell lung cancer
SUV: standard uptake value
TPS: tumor proportion score
KD: Conceptualization, Investigation, Data curation, Supervision, Visualization, Software, Resources, Formal analysis. SF: Writing—original draft, Writing—review & editing, Formal analysis. FM: Formal analysis, Conceptualization, Project administration. GL: Methodology, Data curation, Formal analysis, Investigation. JZ: Data curation, Investigation, Validity. YQ: Software, Methodology, Validity.
The authors declare that they have no conflicts of interest.
Not applicable for this study, which does not include actual patient data.
The patient who provided the image for Figure 1 consented to contribute this data in a retrospective study approved by Johns Hopkins University’s Ethics Committee and Institutional Review Board. The related authorization information is as follows: Ethic Committee Name: JHU IRB, Approval Code: IRB-1 (retrospective patient data) Approval Date: 3/18/2020.
Consent for publication has been obtained.
To request the datasets for this study, please contact the corresponding author.
Not applicable.
© The Author(s) 2024.
Copyright: © The Author(s) 2024. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Brandon Wilkins ... James Bradley
