Affiliation:
1Chemistry Department, Universidad Nacional de Colombia, Bogotá 111321, Colombia
ORCID: https://orcid.org/0009-0001-2770-250X
Affiliation:
2Curauma Biotechnology Center, Pontificia Universidad Católica de Valparaíso, Valparaíso 2362807, Chile
ORCID: https://orcid.org/0000-0002-7829-0568
Affiliation:
1Chemistry Department, Universidad Nacional de Colombia, Bogotá 111321, Colombia
Email: murquizam@unal.edu.co
ORCID: https://orcid.org/0000-0001-5478-606X
Explor Drug Sci. 2025;3:1008103 DOI: https://doi.org/10.37349/eds.2025.1008103
Received: December 24, 2024 Accepted: February 20, 2025 Published: March 26, 2025
Academic Editor: Cheorl Ho Kim, Sungkyunkwan University, Samsung Advances Institute of Health Science and Technology (SAIHST), Republic of Korea
The article belongs to the special issue Bioactive Peptides: Pioneering Innovations in Latin American Research
Epstein-Barr virus (EBV), human papillomavirus (HPV), and hepatitis C virus (HCV) are significant human pathogens associated with various diseases, employing complex molecular mechanisms for cellular entry and immune evasion. Peptide-based research, using more than 700 synthetic peptides, has deciphered some of the molecular interactions between viral proteins and host cell receptors, offering promising diagnostics and therapeutic strategies. In EBV, binding peptides have been identified: 11382, 11389, and 11416 derived from gp350/220; 11435, 11436, and 11438 from gp85 [glycoprotein H (gH)]; and 11521 from BNRF1/p140. Most of these peptide sequences are surface-exposed and are part of the contact regions with human cell receptors, making them promising candidates for strategies aimed at inhibiting EBV invasion of human cells. Peptide 11382 is the target of the neutralizing antibody 72A1; peptides 11382 and 11416 induce interleukin-6 production; peptide 11435 binds to integrin αvβ6, and peptide 11438 triggers a cytokine storm. In the HPV L1 protein, a major component of the viral capsid, peptides 18283 and 18294 have been identified as epithelial cell-binding peptides located on the virus surface. Parts of the sequences are recognized by anti-HPV neutralizing antibodies. These two peptides, along with peptide 18301, have been identified as potential biomarkers for HPV infection because they are recognized by antibodies elicited during natural HPV infection, making them suitable targets for serological detection. In the envelope proteins E1 and E2 from HCV, five hepatocyte- and CD81-positive cell-binding peptides have been identified. The sequences of these peptides contain linear B-cell epitopes recognized by neutralizing antibodies, and some of them have been used to develop serological tests for determining HCV infection. Peptide-based approaches can lead to innovative strategies for the prevention, diagnosis, and treatment of these viral diseases. Additionally, these peptides and their sequences can be used to modulate the immune response and generate tools for cancer theragnostic.
The identification of viral protein regions that bind to human host cell proteins, facilitating viral infection, is critical for understanding the molecular mechanisms of infection and pathogenesis [1]. In this context, synthetic peptides have emerged as powerful tools [2–4]. These peptides can be used to pinpoint pathogen protein regions involved in key biological processes, such as host cell invasion and immune evasion [5]. Additionally, these binding peptides could be used for developing novel strategies for disease detection and treatment.
This review highlights studies from Latin America that explore the use of synthetic peptides to identify protein regions implicated in the invasion of human host cells by Epstein-Barr virus (EBV), human papillomavirus (HPV), and hepatitis C virus (HCV). These investigations shed light on the molecular interactions driving pathogenesis and propose new avenues for therapeutic intervention.
HCV, EBV, and HPV are infectious agents that pose significant global public health challenges due to their high prevalence [6–8], capacity to cause chronic infections, and strong association with the development of various cancers [9, 10]. These pathogens contribute not only to individual morbidity and mortality but also to substantial healthcare and economic burdens worldwide [11–13]. In recent years, significant progress has been made in understanding how viruses recognize and invade host cells, particularly through molecular interactions between viral and host cell proteins. Research using synthetic peptides has been instrumental in identifying specific protein regions involved in these interactions [14]. By pinpointing these critical fragments, strategies can be devised to disrupt these interactions, effectively inhibiting infection. These approaches include developing neutralizing antibodies and other therapeutic interventions aimed at blocking key binding sites, offering promising avenues for infection prevention and treatment [15–17].
EBV, a gamma herpesvirus, is one of the most prevalent human viruses, persisting in over 95% of the global population. Primary EBV infection typically occurs in childhood and is often asymptomatic; however, in adolescence and adulthood, EBV is the cause of infectious mononucleosis (IM), affecting up to 70% of young individuals in developed countries [18]. EBV was the first human virus identified to be oncogenic and is implicated in a range of malignancies accounting for over 200,000 new cases globally each year and around 150,000 deaths [19–21].
Similarly, high-risk HPV (HR-HPV) infection is a significant global public health problem because HR-HPVs have been found in 99.7% of the cervical cancer cases, the second most prevalent among women worldwide and the leading cause of cancer-related mortality mostly, in many developing countries [22]. HR-HPVs are also associated with development of oropharyngeal carcinoma, in fact around 70% of oropharyngeal cancers are HPV16 positive; HR-HPVs have been found in about 90% of anal cancers, 50–60% of penile squamous cell carcinomas, 40–50% of vulvar cancers and around 25% of vaginal cancer, being HPV16 type the most frequently detected in these malignancies. Some studies report that gastric cancers may be associated with EBV or HR-HPV [23–26].
HCV is associated with several malignancies, primarily due to its role in chronic liver disease and the development of between 70–80% of hepatocellular carcinoma (HCC), affecting an estimated 71 million people worldwide [27, 28]. HCV remains a significant global health concern due to its association with chronic liver disease, including cirrhosis and HCC, and is a major agent of parenteral transmission transmitted via parenteral routes [29]. HCV is a major risk factor for HCC, particularly in individuals with chronic hepatitis C infection. Chronic HCV infection is associated with an increased risk of certain non-Hodgkin lymphoma (NHL). HCV may contribute to the development of B-cell lymphoproliferative disorders and an increased risk of skin cancers, including squamous cell carcinoma and melanoma due to immune dysregulation. Moreover, individuals co-infected with HIV and HCV are at an even greater risk for developing liver-related complications and malignancies.
EBV is a widely distributed human herpesvirus that primarily infects B-lymphocytes and epithelial cells [30]. This virus is associated with IM, nasopharyngeal carcinoma and Burkitt’s lymphoma, a highly aggressive B-cell lymphoma, particularly prevalent in Africa [18, 31]; EBV is found in approximately 40–50% of HL cases [32, 33], in about 10% of all gastric cancer cases globally [34], in diffuse large B-cell lymphoma and T-cell lymphomas, particularly in immunocompromised individuals such as post-transplant lymphoproliferative disorder [35, 36]. EBV has been implicated in the pathogenesis of several autoimmune disorders, including systemic lupus erythematosus (SLE), multiple sclerosis (MS), and rheumatoid arthritis (RA) [37, 38].
In 2020, EBV-related cancers accounted for between 239,700 and 357,900 new cases worldwide [39]. EBV-related cancers represent about 1–2% of all human tumors globally, equating to approximately 300,000 new cases annually [21]. Additionally, EBV contributes to around 200,000 cancer-related deaths each year [39].
EBV exhibits tropism for multiple human cell types, including immune and epithelial cells. However, it does not infect these cell types simultaneously because the mechanisms of infection differ between them [40]. The molecular basis underlying the switch between these tropisms remains only partially understood, but a key factor in these processes involves interactions between EBV-encoded proteins and host cell proteins (Figure 1) [41].
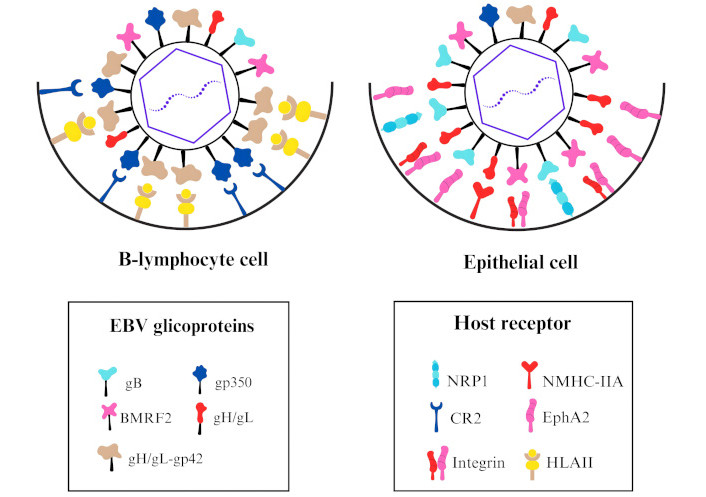
Model illustrating the interaction of the Epstein-Barr virus (EBV) glycoproteins and the host receptors of the target cell. In the B-lymphocyte cell, glycoprotein gp350 interacts with complement receptor type 2 (CR2) for viral attachment, and the complex glycoprotein H (gH)/gL-gp42 interacts with human leukocyte antigens class II (HLAII) for viral binding and promotes the membrane fusion. In the epithelial cell, BMRF2 and gH/gL interact with integrins for viral attachment. For viral binding and the promotion of membrane fusion, gB interacts with neuropilin-1 (NRP1) and ephrin type-A receptor 2 (EphA2), gH/gL interacts with EphA2 and non-muscle myosin heavy-chain IIA (NMHC-IIA) [40, 42]
The infection of epithelial cells involves a multistep process that ensures the successful delivery of the viral genome into the host cell nucleus [42]. EBV utilizes a conserved core fusion machinery comprising glycoprotein B (gB) and the gH/gL complex, which are essential for membrane fusion in both B-cells and epithelial cells [43]. gB, a class III viral fusogen, undergoes conformational changes to mediate membrane fusion. The gH/gL complex acts as a regulator, activating gB upon binding to host cell receptors [44, 45]. The ability of EBV to infect either B-lymphocytes or epithelial cells depends heavily on the composition of the viral envelope proteins, particularly the balance between the epithelial cell-tropic gH/gL complex and the B-cell-tropic gH/gL-gp42 complex [45, 46].
The EBV gH, also known as glycoprotein gp85, is a structural and functional component of EBV’s viral envelope complex, playing also a key role in the invasion of human host cells. This protein facilitates membrane fusion between EBV and human host cells, a critical step for the virus’s entry into B-lymphocytes and epithelial cells [31]. gH (previously referred to as gp85) forms two distinct complexes: a heterodimer (gp85-gp25, also known as gH/gL) shown in Figure 2A, and a heterotrimer (gp85-gp25-gp42, or gH/gL-gp42) shown in Figure 2B [47]. Each complex contributes to the virus’s ability to switch tropism between B-lymphocytes and epithelial cells [48]. The heterodimer primarily mediates infection of epithelial cells by interacting with integrins αvβ6 and αvβ8, while the heterotrimer targets B-lymphocytes through interactions with major histocompatibility complex (MHC) class II molecules [49, 50]. Structurally, gp85 is a glycosylated transmembrane protein with a structure that includes key domains, some of which have been shown to adopt an α-helical configuration, critical for its binding and fusion activity [51].
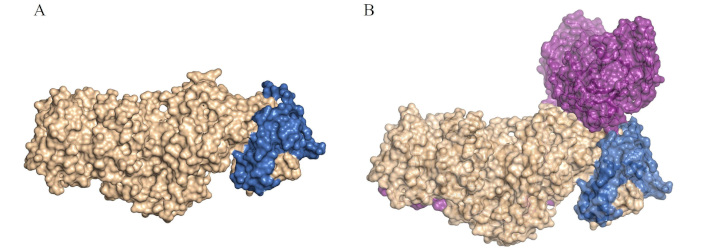
Structural comparison of Epstein-Barr virus (EBV) glycoprotein H (gH)/gL and gH/gL-gp42 complexes in cell tropism. A. Structure of the EBV gH/gL (was adapted from RCSB PDB, PDB ID: 7D5Z [52]) visualized in surface by PyMOL [53]. B. Structure of the EBV gH/gL-gp42 (was adapted from RCSB PDB, PDB ID: 8KHR [54]) visualized in surface by PyMOL [53]. Envelope gH is shown in gold, envelope gL in blue, and protein gp42 in purple. The complexes gH/gL and gH/gL-gp42 are critical for cell tropism. The gH/gL complex facilitates EBV infection of epithelial cells, while the gH/gL-gp42 complex is essential for infecting B-cells [40, 46]
The glycoprotein gp350/220 of EBV is a critical component of the molecular machinery involved in the infection of B-lymphocytes [55, 56] cells, and it elicits neutralizing antibodies preventing virus infections [57]. One EBV gene encodes two major isoforms, gp350 and gp220, which share significant sequence similarity but differ in molecular weight due to alternative splicing [58]. Structurally, gp350 contains a large N-terminal domain that binds to the complement receptor type 2 (CR2/CD21) on B-lymphocytes with high affinity [with a dissociation constant (Kd), in the nanomolar range], facilitating the attachment of EBV to these cells [59–61]. This specific interaction determines EBV’s tropism for B-lymphocytes and plays a pivotal role in the viral life cycle by inducing the synthesis of interleukin-6 (IL-6) [62]. IL-6 is a cytokine that regulates various hematopoietic and immune functions, including B-cell maturation, antibody production, and the extension of B-cell lifespan [63]. Beyond these roles, IL-6 facilitates communication between the innate and adaptive immune systems by modulating the activity of T cells, hepatocytes, and hematopoietic progenitor cells. In the context of EBV infection, IL-6 not only supports the growth of EBV-immortalized B-cells through autocrine and paracrine mechanisms but also contributes to activating intracellular signaling pathways, such as STAT3, which are crucial for the progression of EBV-associated diseases [64].
EBV infects over 90% of adults worldwide and establishes a life-long latency in B-cells [65]. In HL, EBV infection induces a type II latency program, characterized by the expression of latent membrane protein 1 (LMP1) [66, 67]. LMP1 is an oncoprotein that mimics CD40 signaling, activating the nuclear factor kappa-light-chain-enhancer of activated B-cells (NF-κB) pathway, and conferring resistance to apoptosis. LMP1 is also implicated in IL-6 production, reinforcing the link between EBV infection and IL-6-mediated immune modulation [68, 69].
While EBV primarily infects B-lymphocytes and epithelial cells through the receptor-ligand interactions mentioned above, compelling evidence suggests that these interactions are not strictly essential [70]. Other EBV proteins may also play a role in determining host cell tropism. BNRF1/p140 may potentially play a role in alternative EBV invasion pathways of human host cells [71]. As a viral tegument protein, BNRF1 is crucial for the early transcription of EBV genes, ensuring the persistence of the viral genome in the host and allowing it to hijack cellular antiviral mechanisms and evade host immune responses [72, 73]. Evidence suggests that certain regions of p140 might be accessible on the viral surface and antibodies targeting p140 can inhibit infection in B-lymphocytes [74]. BNRF1/p140 is a relatively conserved protein within the EBV tegument layer and is structurally known to reside primarily beneath the viral lipid envelope [75, 76].
Developing effective therapeutic strategies against EBV requires a deep understanding of the molecular interactions that facilitate viral entry into host cells. Extensive studies using synthetic peptides have identified specific regions within these proteins that are crucial for host cell binding and viral entry [77–80]. Additionally, structural analyses of these glycoproteins in complex with their receptors have provided valuable insights for developing novel antiviral strategies, including vaccines and therapeutic antibodies.
Thirty-six synthetic peptides derived from EBV gH, spanning the entire amino acid sequence, have been utilized to identify regions of the protein involved in its interaction with human host cells. This analysis revealed at least three distinct regions of gH, represented by peptides 11435, 11436, and 11438, which exhibit high binding affinity to human cells. These peptides demonstrated significant binding to Raji cells, an EBV-transformed B-cell line, as well as to HeLa cells [51]. These regions were identified before the three-dimensional structure of the protein was determined. Interestingly, most of the sequences of these peptides are exposed in the native structure (Figure 3). These three peptides, or the antibodies induced by immunizing rabbits with them (which recognize live EBV-infected cells), effectively inhibit EBV invasion of cord blood lymphocytes (CBLs). The sequence of peptide 11435 adopts a hairpin structure in its native conformation and binds to integrins αvβ6 and αvβ8 through its KGD motif. This sequence has been extensively studied, with critical residues for interaction with integrins identified. Numerous studies have utilized this peptide, including the development of chimeric peptides designed to induce specific apoptosis in cancer cell lines expressing integrin αvβ6 [81]. In contrast, peptide 11438 adopts an α-helical structure and has been shown to induce a pro-inflammatory cytokine storm [82].
Synthetic analog peptides derived from peptide 11438, designed to adopt a stable α-helix structure as demonstrated by circular dichroism, inhibit EBV invasion of peripheral blood mononuclear cells (PBMCs). These analogs exhibit higher reactivity with anti-EBV antibodies and induce antibodies with greater reactivity against EBV, thereby more effectively inhibiting EBV invasion of epithelial cells compared to the native peptide 11438 or its antibodies. One such analog, peptide 33210, displays biological activity similar to that of EBV-gH proteins. Peptide 33210 binds to normal T-lymphocytes and Raji cells, induces apoptosis in monocytes and Raji cells, but spares normal T cells. Furthermore, peptide 33210 inhibits the differentiation of dendritic cells from monocytes [82].
A promising therapeutic strategy against EBV infection involves the development of nanoparticle-based vaccines targeting the gH/gL and gH/gL-gp42 that elicit potent neutralizing antibodies that effectively inhibit infection of both B-cells and epithelial cells [84, 85]. These vaccines have been shown to generate strong immune responses in animal models, surpassing the levels of naturally acquired immunity in infected individuals. The inclusion of gp42 enhances B-cell-neutralizing antibody titers while maintaining epithelial-cell protection, making it a valuable addition to EBV vaccine formulations [86]. Another approach involves the monoclonal antibody AMMO1, which has been shown to effectively neutralize EBV infection in both B-cells and epithelial cells [87]. Mechanistic analyses suggest that AMMO1 exerts its inhibitory effect by blocking viral fusion with host cells [88].
As previously mentioned, the gB plays a critical role in viral entry and fusion with host cells. Anti-gB antibodies have been identified as the primary neutralizing activity against human cytomegalovirus (HCMV) and EBV infection [85, 89]. In herpesviruses, gH plays a crucial role in membrane fusion, with specific α-helical and coiled-coil domains essential for function [90].
Understanding the molecular details of how gp350/220 interacts with its receptors on B-lymphocytes is essential for designing strategies to block this interaction, thereby preventing B-lymphocyte infection [91]. Such knowledge is critical for developing targeted anti-EBV therapeutics aimed at disrupting these key molecular interactions.
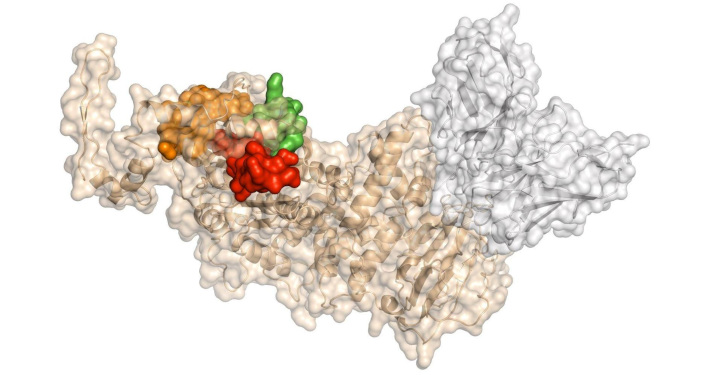
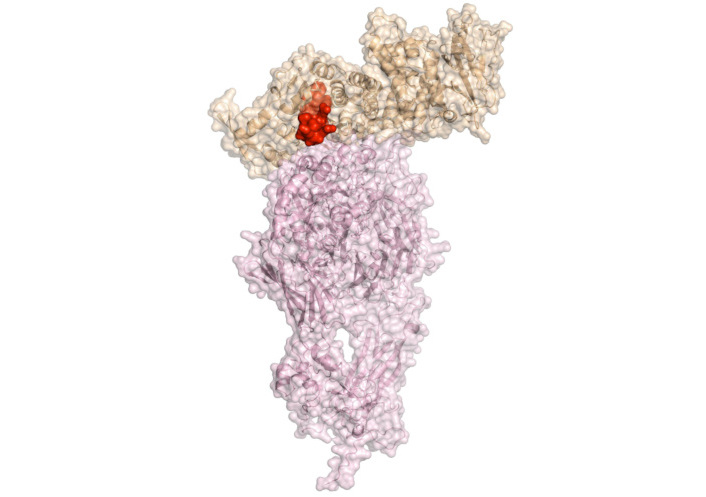
Prior to the determination of the gp350/220 structure and its complex with CD21, Urquiza et al. [92] conducted a detailed analysis to identify regions of gp350/220 involved in B-lymphocyte interaction. This analysis utilized synthetic peptides spanning the entire protein sequence. Through a binding screening of 46 peptides covering the entire gp350’ sequence with B-lymphocytes, they identified three key binding regions. Interestingly, two of these regions were later confirmed to reside within the contact area of gp350/220 with CD21, as revealed by the resolved crystal structure of the gp350/220-CD21 complex (Figure 4). Peptides 11382 and 11389 exhibit sequence homologies with C3dg, one of the human ligands for CR2. This homology suggests that gp350/220 evolved to utilize CR2 on B-cells by mimicking the native CR2-ligand to facilitate cell invasion. The binding of peptides 11382, 11389, and 11416 is specific to cells expressing CR2 protein. Additionally, these peptides inhibit EBV invasion of CBLs, bind to the neutralizing mAb 72A1 (an antibody that prevents EBV from binding to B-lymphocytes), and block mAb 72A1 interaction with EBV [93].
Moreover, peptides 11382 and 11416 induce the expression of IL-6 protein in peripheral blood lymphocytes (PBLs), mimicking the effect of gp350/220 binding to PBLs. This IL-6 induction is inhibited by mAb 72A1, which blocks the activity of both gp350/220 and these peptides. Interestingly, antibodies raised against peptides 11382, 11389, and 11416 effectively inhibit EBV binding and invasion of both PBLs and CBLs [92].
Peptide 11389 binds to PBMCs, inhibiting EBV binding and invasion while inducing IL-6 expression in these cells. Immunization of rabbits with this peptide generates antibodies that recognize EBV-infected cells and inhibit both EBV infection and IL-6 production in PBMCs. Variants of peptide 11389 have been designed with sequence modifications aimed at stabilizing its native β-turn structure by restricting conformational flexibility (reducing the peptide’s configurational entropy) and diminishing non-relevant intermolecular interactions (reducing hydrophobicity). These modifications were carefully implemented to preserve the CD21-interacting residues and maintain the native structure displayed by the gp350 protein. Several of the thirteen synthetic analog peptides exhibit enhanced biological activity compared to peptide 11389, including greater potency in inducing IL-6 expression, inhibiting EBV invasion of PBMCs, interacting with anti-EBV antibodies, and inducing cross-reactive antibodies that recognize EBV [95].
The sequences of peptides 11382 and 11389 have been extensively studied and cited in the literature, encompassing a range of experimental approaches, including molecular dynamics simulations [96], biochemical characterizations [97, 98], and preclinical and clinical trials [99, 100]. These studies have significantly contributed to the rational design of epitope-based peptide vaccines targeting EBV [18, 101]. The sequences of these EBV peptides are being utilized in the design of an epitope-based vaccine, considering that they are often conserved B-cell epitopes located in solvent-accessible regions within the tridimensional structure. The selected epitopes include those derived from EBV envelope glycoproteins, with the aim of eliciting a broad immune response across the population by targeting multiple conserved epitopes [102]. Furthermore, these peptides have been utilized in the development of monoclonal antibodies [103], offering substantial advancements in antiviral therapies and immunological diagnostic tools for EBV infections.
Several vaccines are being developed targeting the gp350 protein region, where these peptides are located. For instance, a novel subunit vaccine has been designed using a Newcastle disease virus (NDV)-based virus-like particle (VLP) platform, which incorporates a chimeric EBV gp350/220-F fusion protein into NDV-derived VLPs. This approach effectively mimics the native EBV structure and facilitates binding to CD21/CD35. Immunization in mice induced robust and long-lasting neutralizing antibody responses, highlighting its potential for EBV vaccine development [104]. Another strategy was a nanoparticle-based vaccine that elicited potent neutralizing antibodies in mice and non-human primates, significantly enhancing immune responses [105].
Remarkably, a vaccine candidate using chimeric VLPs based on the hepatitis B core antigen (HBc149) was designed. This candidate incorporates combinations of three peptides from the receptor-binding domain of EBV gp350, including the 11382 and 11389 sequences. When tested in BALB/c mice, it induced high serum titers and generated neutralizing antibodies that effectively blocked EBV infection in vitro [106]. Moreover, immunization with gp350, combined with other EBV proteins (gH/gL and gB), generates not only humoral immunity (such as the production of neutralizing antibodies) but also cell-mediated immunity. This demonstrates its potential for therapeutic applications to protect against symptomatic infections and associated malignancies [107–109].
Building on the potential of peptide-based strategies for antiviral intervention, sixty-six synthetic peptides covering the complete p140 amino acid sequence have been tested in human cell binding assays, leading to the discovery that peptides 11465 and 11521 specifically bind to EBV-susceptible cell lines, such as Raji, Ramos, and P3HR-1, but not to non-susceptible cells like erythrocytes (Figure 5). Antibodies produced by rabbit immunization with peptide 11521 effectively inhibited EBV binding and invasion of B-lymphocytes [110]. The mechanism by which peptide 11521 or the antibodies induced by it block EBV invasion of host cells remains to be elucidated.
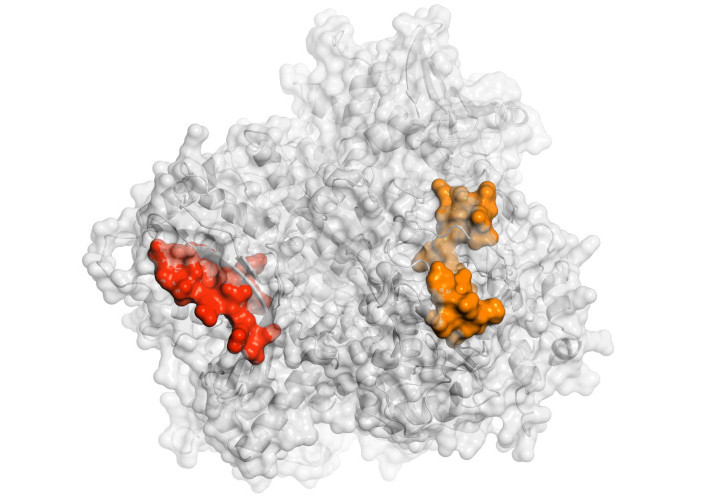
Structure of the Epstein-Barr virus tegument protein BNRF1/p140 modeled by AlphaFold 3 and visualized in surface by PyMOL [53]. The sequence of 11465 and 11521 peptides are shown in red and orange, respectively
In summary, using more than 200 peptides derived from EBV, several peptides from critical regions of EBV proteins are involved in the invasion of human host cells (Table 1). These peptides, identified in Latin America, align with subsequent studies conducted for the therapeutic development of diseases associated with this virus. Most of these peptides are recognized by neutralizing antibodies induced by the complete virus. These peptides inhibit EBV invasion of human host cells, and immunization with them induces neutralizing antibodies. Furthermore, some of these peptides mimic the biological function or phenotypic effects of the full protein.
Sequences of the peptides synthetized for each protein
| Protein | Peptide number | Sequence | Peptide length (amino acids) |
|---|---|---|---|
| Gp350/220 | 11382 | 142HHAEMQNPVYLIPETVPYIK161 | 20 |
| 11389 | 282YVFYSGNGPKASGGDYCIQS301 | 20 | |
| Gp85 [glycoprotein H (gH)] | 11416 | 822PSTSSKLRPRWTFTSPPVTTY841 | 21 |
| 11435 | 181TYKRVTEKGDEHVLSLVFGK200 | 20 | |
| 11436 | 201TKDLPDLRGPFSYPSLTSAQ220 | 20 | |
| 11438 | 241YFVPNLKDMFSRAVTMTAAS260 | 20 | |
| BNRF1/p140 | 11465 | 101GTESALEASGNNYVYAYGPD120 | 20 |
| 11521 | 1221YVLQNAHQIACHFHSNGTDA1240 | 20 |
The ability of integrins to mediate EBV attachment and fusion with epithelial cells underscores their crucial role in viral entry and pathogenesis [111, 112]. This process occurs through their interaction with the ‘KGD’ motif exposed in the gH/gL domain-II [113, 114]. Structural models suggest that this interaction is essential for EBV tropism, as docking studies indicate the formation of a plausible integrin-gH/gL receptor complex (Figure 6) [115]. Additionally, the capacity of gp42 to regulate EBV tropism by potentially obstructing integrin access to the ‘KGD’ (Figure 7) motif further highlights the pivotal role of integrins in viral attachment and fusion [113, 116]. These peptides hold potential for controlling EBV invasion of human cells, given the high prevalence of EBV and its association with malignancies.
Integrins αvβ6 and αvβ8 are not only implicated in EBV-mediated cell fusion but are also upregulated during processes such as tissue remodeling, inflammation, and carcinogenesis [123]. For this reason, peptides that bind with high affinity to these integrins are powerful tools for the development of diagnostic and therapeutic approaches for carcinomas expressing these integrins [81].
Several recombinant gB subunit vaccines, particularly those incorporating the MF59 adjuvant, have been evaluated in clinical trials [77]. These studies confirm that antibodies targeting gB are present in seropositive individuals and contribute to viral neutralization. The most notable clinical result of the gB/MF59 vaccine was its 50% protection efficacy against primary CMV infection in seronegative women of reproductive age [78].
Variations in receptor interactions with viral elements, such as EBV gp350/220, have been implicated in autoimmune conditions, indicating a complex relationship relevant to antibody-mediated responses [79]. Furthermore, the detection of antibodies against specific EBV antigens, which may share sequences or structural motifs with the query, enhances diagnostic sensitivity for conditions like nasopharyngeal carcinoma [80].
Likewise, the impact of single amino acid substitutions on peptide-receptor interactions is particularly evident in viral entry and membrane fusion. In herpesviruses, gH plays a crucial role in membrane fusion, with specific α-helical and coiled-coil domains essential for function [90, 124, 125]. In EBV, mutations in gH residues 52–79 disrupt fusion with both B-cells and epithelial cells [126, 127]. Targeted mutagenesis has shown that even minor changes can fine-tune viral tropism. For instance, R607A and K610A had reciprocal effects on B-cell and epithelial cell fusion, while V592A and R597A impaired only B-cell fusion [128], and the E595A mutation was particularly notable, enhancing B-cell fusion beyond wild-type levels and revealing a direct gp42-gH interaction in the absence of gL [129].
From a translational perspective, studying mutated peptides can aid in the design of more effective peptide-based diagnostics and therapeutics. By understanding the structural and functional implications of antigenic peptide mutations, researchers can refine therapeutic strategies to enhance specificity and minimize off-target effects. Additionally, peptide mutation analysis informs the design of more precise diagnostic tools, improving early detection and disease management [130, 131].
HR-HPV, a DNA virus, plays a pivotal role in the development of cervical cancer and other carcinomas [132]. The World Health Organization recognizes HPV as the primary etiological agent in cervical cancer, with HR-HPV types, including HPV16, HPV18, HPV31, HPV33, and HPV58, being particularly associated with cancer progression [133].
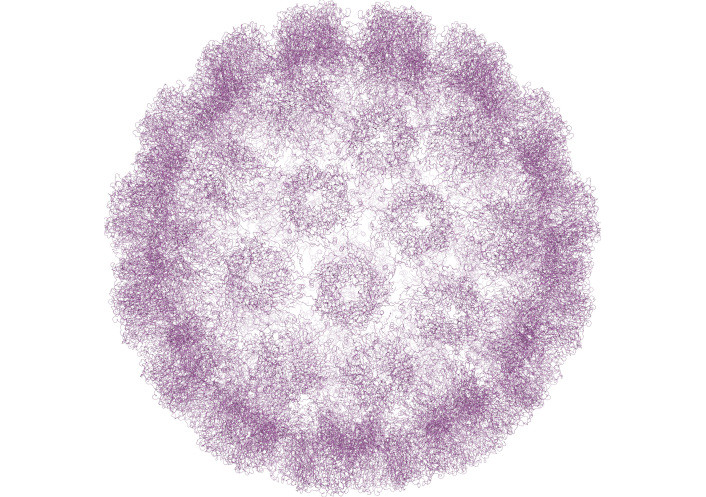
Structurally, HPV is a non-enveloped virus with an icosahedral capsid approximately 55 nM in diameter (Figure 8). The capsid is primarily composed of L1, the major capsid protein, and L2, the minor capsid protein, which encloses the packaged viral DNA. The virion surface is predominantly made up of L1 protein, which contains both variable and conserved regions crucial for immune evasion [134, 135]. The L1 protein has the unique ability to self-assemble into VLPs, typically composed of 72 pentamers of L1 protein. This property makes L1 a potent immunogen, not only during natural persistent HPV infections but also in vaccine immunization strategies [136].
HPV infects basal epithelial cells (Figure 9) through micro abrasions that facilitate the virus’s interaction with human host cells [139]. During this process, the HPV L1 protein initially binds to heparan sulfate proteoglycans (HSPGs) on the host cell surface, likely through the positively charged regions located on the L1 pentamer surface [140]. This interaction is a critical step for viral attachment. Subsequently, the L1 and L2 proteins interact with additional cellular receptors, such as epidermal growth factor receptors (EGFRs) and fibroblast growth factor receptors (FGFRs), to facilitate viral entry [141]. Upon internalization, the viral genome is transported to the host nucleus, where it can integrate into the host DNA [142]. Two viral proteins, E6 and E7, play central roles in cancer development. E6 specifically targets p53 for ubiquitination, leading to its degradation. Additionally, E6 is involved in the induction of epithelial-mesenchymal transition (EMT), a key step in cancer progression [143, 144]. E7, on the other hand, binds to the retinoblastoma protein (pRb) and interferes with cyclin-dependent kinase inhibitors such as p21 and p27, thereby promoting cell cycle progression [145, 146]. Both p53 and pRb are essential for regulating the cell cycle and maintaining genomic integrity [147, 148]. Their inactivation by E6 and E7 disrupts these processes, leading to uncontrolled cell proliferation and malignancy.
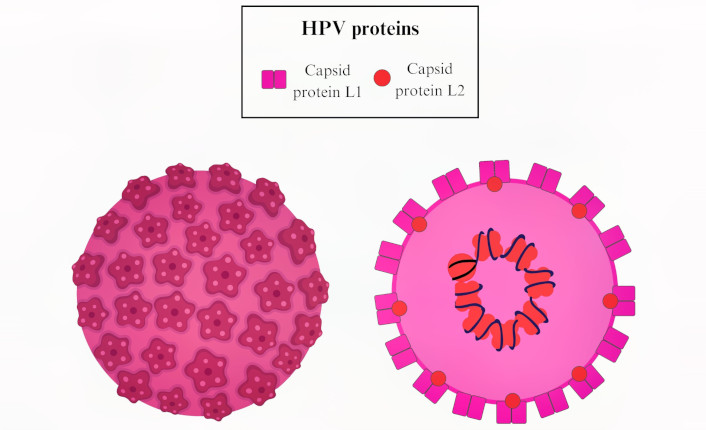
Model illustrating the human papillomavirus (HPV) structure with the capsid protein L1 and L2. HPV infects basal epithelial cells through micro abrasions that facilitate the virus’s interaction using L1 and L2 proteins to bind different receptors on human epithelial host cells
Integrins serve as secondary receptors that may assist in the internalization process. There is evidence indicating that binding correlates with the levels of α6 integrin, and the signaling pathways are important for HPV16 infection [149]. Annexin A2 has been identified as a specific receptor for the L2 minor capsid protein. The proposed model emphasizes that multiple receptors and coreceptors can coalesce into a receptor complex that includes HSPGs, integrins, CyPB, tetraspanin, GFRs, and A2t. Overall, the interaction between L1 and cell receptors is characterized by initial binding to HSPGs, subsequent conformational changes, signaling pathway activation, and eventual engagement with other receptors like integrins and annexin A2, illustrating a complex and multistep molecular process essential for viral entry [150].
L1-binding regions to VERO and HeLa cells have been identified using fifty-six synthetic 20-mer peptides derived from the complete amino acid sequences of the HPV16 and HPV18 L1 proteins. Peptides 18283 (residues 54–77) and 18294 (residues 274–308) from HPV16 L1, as well as peptides 18312 (residues 59–78) and 18322 (residues 259–278) from HPV18 L1, contain these binding regions. The sequence PKVSGLQYRVFR is conserved in the L1 proteins of HPV16 and HPV18 and is present in the binding peptides 18283 and 18312. Similarly, the sequence LYIKG is conserved in both HPV16 and HPV18 and is found in binding peptides 18294 and 18322.
According to the crystal structure of the HPV16 L1 protein [151], the amino acid sequences of these peptides, exposed to the solvent on the surface of the VLP pentamer, correspond to PNNNKILVP and LYIKGSGSTAN, which are likely regions involved in cell interaction. The affinity constants for cell binding of peptides 18283 and 18294 are 300 nM and 600 nM, respectively. While these affinities are moderate, considering that these peptides are repeated at least five times on the pentamer, the effective binding affinity could be significantly enhanced due to potential positive cooperativity during cell invasion. The receptors for these peptides appear to be cell surface proteins sensitive to trypsin and chymotrypsin, with molecular weights of approximately 69 and 54 kDa. Importantly, peptides 18283 and 18294 specifically inhibited HPV16 VLP binding to HeLa cells. Based on the reported structures of L1 and VLPs, both peptide sequences are located close to one another on the VLP surface, forming outer binding sites with potential receptor interaction regions. Furthermore, it has been reported that a conserved motif within peptide 18294 serves as a target for neutralizing antibodies [152].
Interestingly, peptides 18283, 18294, and 18301 (Table 2) are recognized by antibodies generated during natural persistent infections with HR-HPVs, which are strongly associated with cervical lesions and an increased risk of cancer. In the pentameric structure of the HPV16 L1 protein, shown in Figure 10, it is evident that these peptides are surface-exposed which increases the likelihood that these will act as immunodominant epitopes, eliciting a strong immune response. These peptides have demonstrated high sensitivity and specificity in distinguishing between women with HPV-associated cervical lesions and those with normal cytology. For instance, peptides 18283 and 18294 exhibited sensitivities of 92–97% and specificities of 89–95% for detecting precancerous lesions, with minimal reactivity observed in sera from women with normal cytology [153]. These findings highlight the potential of L1-derived peptides as biomarkers for detecting women with persistent HR-HPV infections in large-scale screening programs.
Sequences of the peptides synthetized from the L1 protein
| Protein | Peptide number | Sequence | Peptide length (amino acids) |
|---|---|---|---|
| L1 | 18283 | 55PNNNKILVPKVSGLQYRVFR74 | 20 |
| 18294 | 275LYIKGSGSTANLASSNYFPT294 | 20 | |
| 18301 | 415EDTYRFVTSQAIACQKHTPPA435 | 21 | |
| 18322 | 259FWNRAGTMGDTVPQSLYIKG278 | 20 | |
| 18312 | 59QDIPKVSAYQYRVFRVQLPD78 | 20 |
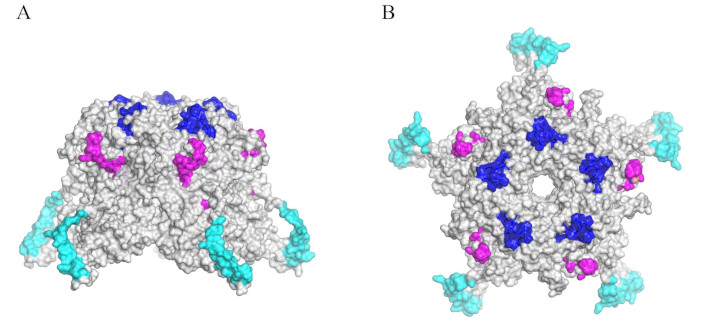
Pentamer structure of human papillomavirus 16 (HPV16) L1 (was adapted from RCSB PDB, PDB ID: 1DZL) visualized in surface by PyMOL [53]. The sequence of 18294, 18283, and 18301 peptides are shown in blue, magenta, and cyan, respectively. A. The structure is shown in side view. B. The structure is shown in front view
Synthetic peptides 18283, 18294, and 18301 displayed comparable sensitivity (75–83%) and higher specificity than VLPs for detecting women with HR-HPV infections in a study involving 391 HPV-infected female volunteers. Notably, the non-variable C-terminal L1 peptide 18301 demonstrated the ability to detect protective antibodies in HPV DNA-negative women with atypical squamous cells of undetermined significance (ASC-US) [154]. This peptide, which is surface-exposed on the L1 protein, is specifically recognized by antibodies from patients with HPV-associated cervical lesions or cervical cancer. In serological tests, it achieved remarkable sensitivity (88.36%), specificity (99.39%), and overall classification accuracy (94.21%) for detecting HR-HPV cases [155].
It has been claimed that these peptides are valuable for screening programs in resource-limited settings, offering additional utility in monitoring women at higher risk of developing cervical cancer and in distinguishing between HR and non-infected patients. This makes them promising candidates for developing cost-effective and reliable serological screening tools in clinical and resource-limited settings.
Peptides SPINNTKPHEAR and MKIPNNKLFLPV, identified using a phage display random peptide library, expressed on the surface of bacteriophages and screened with antibodies present in the sera of HR-HPV patients. These peptides exhibit sequence similarity to the HPV16 L1 protein, particularly peptide 18283, and have demonstrated potential for early HPV detection [22]. This is especially significant for women with precancerous cervical lesions, representing a critical step toward the development of accessible diagnostic tests in low-resource settings where polymerase chain reaction (PCR)-based tests remain cost-prohibitive.
Prophylactic and therapeutic peptide-based vaccines offer advantages in terms of stability, safety, and ease of production but face challenges such as low immunogenicity and MHC restriction, necessitating the use of adjuvants to enhance immune responses. Despite these challenges, short peptide vaccines, such as CIGB-228, necessitate human leukocyte antigen (HLA) typing but have shown promising results in HPV16-positive CIN patients, inducing T-cell responses and lesion regression [156]. In contrast, synthetic long peptides (SLPs) bypass MHC restriction but rely on antigen-presenting cells (APCs) for processing and their effectiveness in advanced cervical cancer remains under investigation [157, 158].
At least three licensed prophylactic HPV vaccines are currently available: Gardasil® 4 (quadrivalent vaccine)—available since 2006, it protects against four HPV types: 6, 11, 16, and 18; Cervarix™ (bivalent vaccine)—available since 2007, it targets HPV types 16 and 18; Gardasil® 9 (nonavalent vaccine)—available since 2014, it provides protection against nine HPV types: 6, 11, 16, 18, 31, 33, 45, 52, and 58 [157]. These vaccines are based on VLPs composed of the major papillomavirus capsid protein L1, which induces high titers of neutralizing antibodies. On the other hand, therapeutic HPV vaccines include: live vector-based vaccines—these use genetically modified viruses to deliver HPV antigens to the immune system, inducing a response against HPV-infected cells; protein vaccines—composed of the oncoproteins E6 and E7, these vaccines activate the immune system to destroy cells expressing these antigens; nucleic acid vaccines—these utilize DNA or RNA encoding HPV antigens to generate an immune response against HPV infected cells; whole cell vaccines—these involve the use of whole tumor cells that express HPV antigens, eliciting an immune response targeting not only HPV proteins but also other tumor-associated antigens [159].
For example, RNA vaccines are being developed for HPV infection utilizing lipid nanoparticle (LNP) encapsulation to deliver mRNA that encodes chimeric proteins formed by fusing HPV E7 with herpes simplex virus gD, which have demonstrated promising immunogenicity and efficacy in animal models [160]. Preliminary results in animal models have shown promising immunogenicity and efficacy. New topical compounds with cytotoxic effects or those designed to stimulate the immune response are being utilized to eliminate HPV-related lesions. For instance, preparations containing salicylic acid and 5-fluorouracil are common in treating cutaneous warts and other HPV-induced lesions [161]. Additionally, some approaches involve the combination of existing treatments such as cryotherapy, laser therapy, and cytotoxic agents [162].
HCV infection remains a significant global health challenge, with estimates indicating that approximately 200 million people worldwide are infected [163]. HCV is primarily transmitted through parenteral routes, such as blood transfusions, although a substantial percentage of cases involve unknown transmission pathways [164, 165]. Chronic HCV infection carries an HR of progression to severe liver conditions, including cirrhosis and HCC [166].
The structure and replication mechanism of HCV is critical to understanding its pathogenicity and for designing effective detection and therapeutic strategies. HCV is a positive-sense RNA virus that encodes a single large polyprotein of approximately 3,011 amino acids [167]. This polyprotein is subsequently cleaved into structural and non-structural proteins. The structural proteins include the core protein, which forms the viral nucleocapsid, and the envelope proteins E1 and E2, which are essential for viral entry (Figure 11) [168].
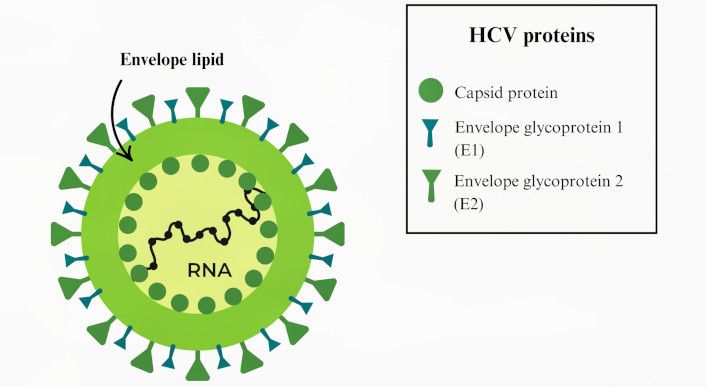
Model illustrating the hepatitis C virus (HCV) structure. HCV is a positive single-stranded RNA virus belonging to the Flaviviridae family; its genome is approximately 9.6 kb in length, that encodes a large polyprotein precursor, which is cleaved into at least 10 distinct proteins essential for both viral replication and modulation of host cell functions [169]
The E1 and E2 envelope proteins facilitate HCV binding to hepatocytes and possibly B-cells [170]. Evidence indicates that CD81, a tetraspanin molecule, is a key binding partner for E2 on the surface of host cells. Interestingly, some studies suggest that HCV can interact with hepatocytes independently of CD81, implying that alternative receptors or co-receptors may contribute to the viral entry process [171]. CD81 and claudin-1 are coreceptors during HCV entry, which is essential for the successful internalization of the virus; in fact, anti-E1 antibodies that modulate this co-receptors association inhibit HCV entry processes [172].
The initial attachment of HCV to host cells involves interactions between E1 and various receptors, including low-density lipoprotein receptors (LDLRs) and glycosaminoglycans (GAGs). This is followed by co-receptor binding to CD36 and scavenger receptor class B type I (SR-BI), which promote viral internalization. On the other hand, HCV E2 interacts with CD81, SR-BI, claudin-1, and occluding to mediate viral entry into host cells [173, 174]. The human SR-BI is also necessary for HCV entry, especially in cells that express CD81, indicating its importance in the infectivity process [175]. Besides CD81 and SR-B1, additional hepatocyte-specific co-factors are likely involved in facilitating viral entry, suggesting a complex interaction necessary for successful infection [176]. According to experimental and clinical evidence, HCV induces immune evasion through mainly high variability of these proteins but there are regions involved in cell receptor interactions that have not high variability being targets for effective therapeutic interventions [177].
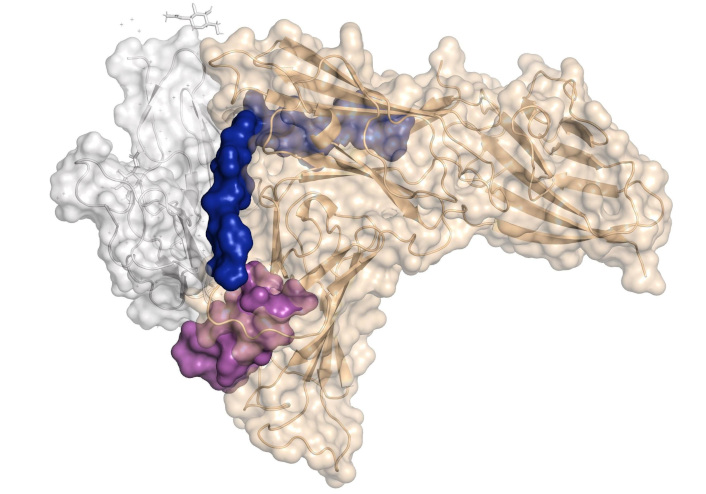
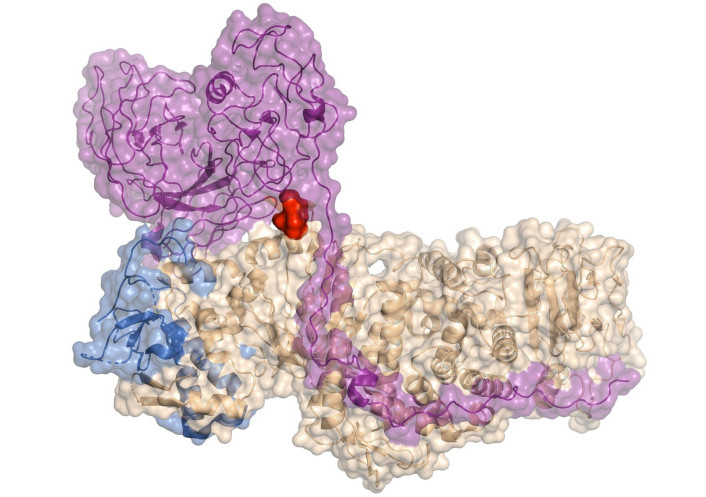
To identify HCV-binding regions within the envelope proteins E1 and E2 that interact with human hepatocytes, 18 synthetic peptides derived from E1 and 34 synthetic peptides from E2 were tested for binding to the human HCC cell line HepG2 [178]. Among these synthetic peptides, twelve exhibited high binding activity to these cells, allowing the identification of six hepatocyte-binding sequences (HBSs). These findings revealed two binding regions in E1, specifically high activity binding peptides (HABPs) 4913 and 4918, and four binding regions in E2, referred to as region I, region II, region III, and region IV. Table 3 presents the sequences of the peptides that are more exposed in the complex, and Figure 12 shows their location within the protein structure.
Sequences of the peptides synthetized from hepatitis C virus (HCV) envelope proteins
| Protein | Peptide number | Sequence | Peptide length (amino acids) |
|---|---|---|---|
| E1 | 4913 | YQVRNSTGLYHVTNDCPNSS | 20 |
| 4918 | MTPTVATRDGKLPATQLRRHY | 21 | |
| E2 | Region I | ETHVTGGSAGHTVSGFVSLLY | 21 |
| Region II | HHKFNSSGCPERLASCRPLT | 20 | |
| Region III | PTYSWGENDTDVFVLNNTR | 19 |
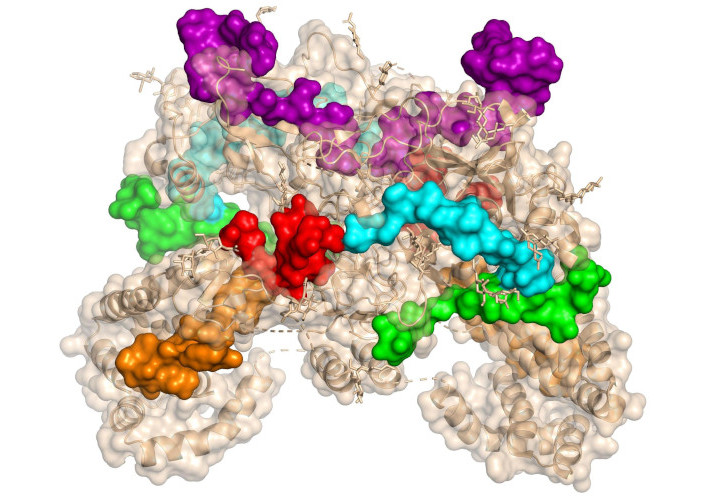
Structure of homodimer complex of hepatitis C virus (HCV) E1/E2 (was adapted from RCSB PDB, PDB ID: 8RJJ) visualized in surface by PyMOL [53]. The sequence of 4913 and 4918 peptides from E1 protein is shown in red and orange, respectively. The sequences of the peptides from the E2’s regions I, II, and III are shown in green, cyan, and magenta, respectively
Within the E1 protein, peptides 4913 and 4918 were identified as essential for binding, while in the E2 protein, peptides spanning regions I-IV facilitated attachment. Several of these HABPs also demonstrated the ability to bind to CD81-positive Raji cells, indicating their potential role in HCV interactions with both liver and immune cells [178]. The specific affinity of these sequences for hepatocyte receptors, along with their location within immunogenic domains, highlights their potential as novel therapeutic targets or vaccine candidates.
The hypervariable region 1 (HVR-1) of the E2 protein is particularly notable for eliciting antibodies capable of inhibiting HCV binding to target cells, marking it as a promising vaccine candidate. However, the high sequence variability of HVR-1, likely driven by immune pressure, complicates its use as a universal target. Identifying additional conserved B-cell epitopes within the E1 and E2 proteins is therefore critical for the development of effective vaccines and therapeutic agents targeting HCV [179].
An immunogenic peptide, YEVRNVSGIYHVTNDCSNSS, has been reported to share high homology with HABP 4913 (YQVRNSTGLYHVTNDCPNSS) located in the N-terminal region of the E1 protein [180]. Similarly, HABP 4931 (part of the E2 protein and underlined) belongs to HVR-1 (ETHVTGGSAGHTVSGFVSLLAPGAKQN) and is predominantly exposed on the viral surface [181]. Evidence suggests that the N-terminal regions of E1 and E2 (where HABPs 4913 and 4931 are located) are surface-exposed [181]. Antibodies targeting HVR-1 have been shown to neutralize the virus [181–183] and confer homologous protection in chimpanzees and rabbits [184–186]. Anti-HVR-1-specific antibodies, particularly at the N-terminal, middle, or C-terminal regions where HABP 4931 is located, can prevent HCV infection [181].
Notably, the N-terminal region of HVR-1 has been reported to inhibit in vitro HCV binding to human fibroblast cells. Furthermore, antibodies from rabbits targeting the C-terminal region of HVR-1 block HCV absorption to cells [187], suggesting that HABP 4931 plays a critical role in HCV binding and cellular invasion. HVR-1 is the major antigenic region recognized by anti-HCV antibodies, containing linear B-cell epitopes [187, 188].
Peptide-1 [189] (TTDRSGAPTYSWGANDTDVFV) has been reported as a linear B-cell epitope and contains part of the sequences of HABPs 4943 (PVYCFTPSPVVVGTTDRSGA) and 4944 (VVVGTTDRSGAPTYSWGEND). Another B-cell epitope (TWGENETDVLLLNNTRPPQ) [190] shares sequence homology with HABP 4944 and peptide 4945 (APTYSWGENDTDVFVLNNTR). The sequences of peptides 4944 and 4945 have been shown to be antigenic, as they are recognized by patients’ sera and are immunogenic, inducing the production of monoclonal antibodies [178]. This peptide is one of the 12 peptides identified from a library of 126 overlapping peptides (15-mers offset by 10 amino acids for E1, by 11 amino acids for E2) that inhibited HCV infection by more than 80% at a concentration of 10 nM. The library covers the envelope protein E1E2 of the HCV JFH1 strain (GenBank no. AB047639) [191]. These findings highlight the potential of peptide-based inhibitors in HCV therapeutic development [192, 193].
Synthetic peptide-based assays for HCV detection have shown significant promise, particularly in low-resource settings, as peptides targeting specific HCV epitopes offer cost-effective alternatives to commercial recombinant protein-based assays. In a study, synthetic peptides derived from structural and non-structural HCV proteins were used to develop two enzyme immunoassays (EIAs): EIA-Spep for initial screening and EIA-Cpep for confirmatory testing [194].
The EIA-Spep screening assay, which features a mixture of three peptides, demonstrated exceptional performance, achieving a specificity of 99.9% and a negative predictive value of 100% when tested on blood donor samples from Central American countries [195]. Notably, the confirmatory EIA-Cpep assay provided more definitive results, significantly reducing indeterminate outcomes compared to the Abbott Matrix confirmatory test.
This synthetic peptide-based approach shows great potential for broad and accessible HCV testing in resource-constrained settings. By enabling cost-effective and reliable screening, these assays could facilitate regular blood donor testing and improve the identification of HR populations, ultimately helping to curtail HCV transmission.
Peptide-based research has emerged as a powerful tool not only to understand how EBV, HPV, and HCV infect human cells and interact with the immune system but also to combat these viral infections and the diseases they cause. These viruses are significant contributors to the global disease burden, and peptide-based studies offer promising strategies for diagnostics, therapeutics, and vaccine development.
This review highlights the significant work conducted to understand how these viruses infect human cells using more than 700 synthetic peptides, including analog peptides from six proteins of three different viruses, and in vitro assays with at least ten different human cell lines and human cells. Using these peptides, several binding regions have been identified on the surface of these viruses: seven in three proteins of EBV, two in the L1 protein from HPV, and four in HCV. Most of these regions are recognized by neutralizing antibodies generated either through natural or artificial immunization; additionally, some peptides from these regions are capable of inducing neutralizing antibodies.
In the case of EBV, regions identified in gp350 and gH correspond to binding regions based on the crystal structure of the complex. Some EBV-binding peptides are recognized by neutralizing antibodies, induce IL-6 production, and trigger a cytokine storm. In HPV, the identified binding peptides, including peptide 18301, and in HCV, the binding peptides are recognized by antibodies induced through natural immunization. These peptides have also been used to study the natural history of infection for these viruses.
Peptides derived from the HPV L1 capsid protein show significant potential in cervical cancer screening, particularly in low-resource settings. These peptides exhibit high sensitivity and specificity in detecting HR-HPV infections and distinguishing between patients with cervical lesions and those with normal cytology, offering alternatives or complements to current diagnostic methods.
Similarly, for HCV, synthetic peptide-based assays have demonstrated effectiveness in both initial screening and confirmatory testing, showing remarkable specificity and predictive value. Peptides targeting epitopes within the envelope proteins E1 and E2 have identified HBSs that are critical for viral entry, providing valuable insights into HCV pathogenesis and highlighting potential targets for vaccine development. In brief, these peptides are valuable tools for the detection, treatment, and vaccine development against these viruses and the diseases they cause.
CBLs: cord blood lymphocytes
CR2: complement receptor type 2
EBV: Epstein-Barr virus
EIAs: enzyme immunoassays
EphA2: ephrin type-A receptor 2
gB: glycoprotein B
HABPs: high activity binding peptides
HBSs: hepatocyte-binding sequences
HCC: hepatocellular carcinoma
HCV: hepatitis C virus
HL: Hodgkin lymphoma
HPV: human papillomavirus
HR: high-risk
HSPGs: heparan sulfate proteoglycans
HVR-1: hypervariable region 1
IL-6: interleukin-6
IM: infectious mononucleosis
LMP1: latent membrane protein 1
MHC: major histocompatibility complex
NDV: Newcastle disease virus
PBLs: peripheral blood lymphocytes
PBMCs: peripheral blood mononuclear cells
pRb: retinoblastoma protein
SR-BI: scavenger receptor class B type I
VLP: virus-like particle
The authors would like to express their sincere gratitude to the Universidad Nacional de Colombia for its continuous support and encouragement in fostering scientific research and innovation. We express our deepest gratitude to Prof. Manuel E. Patarroyo, whose invaluable guidance and mentorship were instrumental in shaping the research review described here. Sadly, Prof. Manuel E. Patarroyo passed away on January 9, 2025, during the preparation of this manuscript. His dedication to scientific inquiry and his profound impact on the field will always be remembered. Molecular graphics and analyses performed with PyMOL.
DPJ: Conceptualization, Investigation, Writing—original draft, Writing—review & editing. FGQ: Conceptualization, Investigation. MUM: Conceptualization, Validation, Writing—review & editing, Supervision. All authors read and approved the submitted version.
Fanny Guzmán-Quimbayo who is the Guest Editor of Exploration of Drug Science had no involvement in the decision-making or the review process of this manuscript. The other authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
The dataset analyzed for this study can be found in the RCSB Protein Data Bank RCSB PDB: Homepage.
Not applicable.
© The Author(s) 2025.
Open Exploration maintains a neutral stance on jurisdictional claims in published institutional affiliations and maps. All opinions expressed in this article are the personal views of the author(s) and do not represent the stance of the editorial team or the publisher.
Copyright: © The Author(s) 2025. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
View: 4471
Download: 76
Times Cited: 0
Constanza Cárdenas ... Fanny Guzmán
Andrea Barragán-Cárdenas ... Javier García-Castañeda
Natalia Ardila-Chantré ... Javier Eduardo García-Castañeda
Jésica A. Rodríguez ... Silvia A. Camperi
Lucero Ruiz-Mazón ... Mireya de la Garza
Vaezeh Fathi Vavsari, Saeed Balalaie
Vanesa Herlax
Karen Johanna Cárdenas-Martínez ... Javier Eduardo García-Castañeda
