Affiliation:
Australian Institute of Tropical Health and Medicine, James Cook University, Cairns, QLD 4878, Australia
ORCID: https://orcid.org/0000-0002-1527-4026
Affiliation:
Australian Institute of Tropical Health and Medicine, James Cook University, Cairns, QLD 4878, Australia
ORCID: https://orcid.org/0000-0001-5047-0711
Affiliation:
Australian Institute of Tropical Health and Medicine, James Cook University, Cairns, QLD 4878, Australia
Affiliation:
Australian Institute of Tropical Health and Medicine, James Cook University, Cairns, QLD 4878, Australia
ORCID: https://orcid.org/0000-0002-0896-8441
Affiliation:
Australian Institute of Tropical Health and Medicine, James Cook University, Cairns, QLD 4878, Australia
Email: michael.smout@jcu.edu.au
ORCID: https://orcid.org/0000-0001-6937-0112
Affiliation:
Australian Institute of Tropical Health and Medicine, James Cook University, Cairns, QLD 4878, Australia
Email: norelle.daly@jcu.edu.au
ORCID: https://orcid.org/0000-0002-4697-6602
Explor Drug Sci. 2023;1:172–179 DOI: https://doi.org/10.37349/eds.2023.00012
Received: January 06, 2023 Accepted: April 20, 2023 Published: June 30, 2023
Academic Editor: Xuechen Li, The University of Hong Kong, China
The article belongs to the special issue Exploring Potential Drugs from Natural Products
Aim: Identification of small bioactive regions in proteins and peptides can be useful information in drug design studies. The current study has shown that an inter-cysteine loop of the N-terminal domain of Opisthorchis viverrini granulin-1 (Ov-GRN-1), a granulin protein from the flatworm liver fluke Opisthorchis viverrini which has potent wound healing properties, maintains the bioactivity of the full-length protein.
Methods: Peptides corresponding to the three inter-cysteine loops of the N-terminal domain were produced using synthetic chemistry, and their structures and bioactivities were analyzed using nuclear magnetic resonance (NMR) spectroscopy and cell proliferation assays, respectively.
Results: As expected for such small peptides, NMR analysis indicated that the peptides were poorly structured in solution. However, a seven-residue peptide corresponding to loop 2 (GRN-L2) promoted cell proliferation, in contrast to the other fragments.
Conclusions: The results from the current study suggest that GRN-L2 might be responsible, in part, for the bioactivity of Ov-GRN-1, and might be a useful lead molecule for subsequent wound healing studies.
Identification of small bioactive regions in proteins and peptides can be useful in drug design studies, including peptide mimetic [1, 2] and grafting studies [3–7]. Such bioactive peptides might also be drug leads in their own right, as they are likely to be less immunogenic than larger proteins and easier and cheaper to manufacture [8, 9]. One of the unique approaches to produce bioactive peptides is a process referred to as protein “downsizing” [9], where small, bioactive regions of proteins are produced in isolation. Although downsizing is not as effective where dis-continuous regions are involved in bioactivity, there are several examples where this approach has been effective. For example, downsizing C3a, a 77-residue human inflammatory protein, resulting in the identification of a tripeptide with the same high potency, functional profile, and specificity of action as the full-length C3a protein, but with higher plasma stability and bioavailability [9].
The downsizing approach has also been effectively applied to the granulin protein Opisthorchis viverrini granulin-1 (Ov-GRN-1) from the parasitic human liver fluke, Opisthorchis viverrini. Ov-GRN-1 is a 6 kDa protein that induces angiogenesis and accelerates wound repair [10]. Although Ov-GRN-1 has potent wound-healing properties, low yields in recombinant expression have limited its development as a potential wound-healing agent [11], and prompted studies aimed at producing smaller versions with synthetic methods. A truncated form (GRN12–35_3s) corresponding to residues 12–35 was produced using solid-phase peptide synthesis and folded into a relatively well-defined structure containing a β-hairpin with three disulfide bonds [12]. This truncated form of Ov-GRN-1 had similar potency to the full-length protein in an in vivo wound healing assay [12] and contains two native disulfide bonds and one non-native disulfide bond, based on the full-length granulin disulfide connectivity [13].
GRN12–35_3s is still a relatively large/structurally complex peptide that comprises three inter-cysteine loops. To determine if Ov-GRN-1 can be downsized even further, the three inter-cysteine loops of Ov-GRN12–35_3s, referred to as GRN-L1, GRN-L2, and GRN-L3, were individually synthesized using solid-phase peptide synthesis. Cysteine residues were included at the N- and C-termini to allow the formation of a disulfide bond to bring the termini in close proximity and form a loop structure. The structures and cell proliferation effects of these peptides were examined.
Standard solid-phase peptide synthesis methods were used for the synthesis of the truncated granulin peptides. The peptide chains were assembled on 2-chlorotrityl chloride resin (Peptides International, USA, product code RCT-1056-PI) using manual procedures as previously described [13]. Each peptide contained two cysteine residues, and oxidation of these cysteine residues into a disulfide bond was carried out by incubating the peptides in 100 mmol/L ammonium bicarbonate (pH 8.2) for 24 h at room temperature. The pH was checked after dissolution to confirm the solutions were still at pH 8.0. Preparative reversed-phase high-performance liquid chromatography (RP-HPLC) was used to purify the peptides following the oxidation reaction (Phenomenex Jupiter C18, 10 μm, 300 Å, 250 mm × 21.2 mm), and the purity assessed using analytical RP-HPLC (Phenomenex Jupiter 4 μm Proteo column C12, 10 μm, 90 Å, 150 mm × 2 mm). A 1% gradient was used to monitor the purity (0–60% solvent B over 60 min with a flow rate of 0.4 mL/min) and the absorbance was monitored at 214 nm and 280 nm. A 5800 MALDI TOF/TOF mass spectrometer (SCIEX, Framingham, MA, USA) and an LCMS-2020 (Shimadzu, Japan) were used to analyze the masses of the fractions.
The three-dimensional structures of the peptides were analyzed using nuclear magnetic resonance (NMR) spectroscopy data and calculations based on torsion angle dynamics. Peptides were dissolved at a concentration of ~ 0.2 mmol/L in 90% H2O/10% D2O (v/v), pH 4.5, and 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS; Cambridge Isotope Laboratories) used as an internal chemical shift reference. A 600-MHz AVANCE III NMR spectrometer (Bruker, Karlsruhe, Germany) was used to record spectra which included TOCSY (80 ms mixing time), NOESY (200–300 ms mixing time), and DQF-COSY experiments. The relaxation delay for the experiments was set to 1 s and the temperature was set to 290 Kelvin (K). TOPSPIN software (Bruker, Billerica, MA, USA) was used to process the data, and CCPNMR [14] was used to analyze and assign the spectra based on established protocols [15]. The program CYANA was used to calculate the three-dimensional structures, using a macro for automated assignment of the non-intra-residue NOEs [16]. Restraints for dihedral angles were predicted by TALOS-N and incorporated into the structure calculations [17]. Distance restraints for the disulfide-bonds were included in the calculations. Structures were visualized using MOLMOL, a molecular graphics program for displaying and analyzing the three-dimensional structures of molecules [18].
The cell line used for the cell proliferation experiments (human skin normal fibroblast cell line, 1BR.3.GN), was obtained from the European Collection of Authenticated Cell Cultures (ECACC, product code 90020509). The conditions used to culture and maintain the cells are those previously described [13] (i.e. complete media: Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F12), 1 × GlutaMAX (Thermofisher, product code 10565042) and 1 × antibiotic/antimycotic (penicillin, streptomycin, and amphotericin B; Thermofisher, product code 15240062), 10% fetal bovine serum (FBS; Thermofisher, product code 12664025) at 37°C and 5% CO2). A 21-residue peptide was used as the negative control. Cell proliferation assays were done in the presence of low nutrient media: DMEM/F12 as above with 0.5% FBS.
To monitor the cell proliferation, 1BR.3.GN cells seeded at 5,000 cells per well were grown overnight at 37°C in 150 μL of complete media. Agilent E-plates (Agilent) were used for the assays. The cells were monitored using an xCELLigence SP system (Agilent). This system uses gold microelectrodes, which are integrated into the base of tissue culture plates, to measure electrical impedance. Low-nutrient media was used to wash the cells (three times) and incubate the cells (minimum of 6 h) prior to treatment with peptides. Peptides were quantified based on absorbance measurements at 214 nm and predicted extinction coefficients based on the amino acid compositions [19, 20]. Treatments were prepared at 8.5 × target concentration and added to each well in 20 μL, for a final total volume of 170 μL and 1 × concentration. The pH was checked after dissolution to confirm the solutions were still at pH 7. The cell proliferation experiments were carried out over a period of 5–6 days following treatment with the peptides. The xCELLigence system was used to record cell indexes at intervals of 1 h over this time period. To determine the cell proliferation ratios cell index readings were normalized before peptide treatment.
A Brown-Forsythe test was used to determine variance homogeneity. It was not possible to determine if the data displayed a normal distribution because of the limited number of data points. A one-way ANOVA test with Holm-Sidak’s multiple comparison corrections was used to compare the cell proliferation rates between treatments at each concentration and the nearest peptide control concentration in GraphPad Prism 9.0.
Three Ov-GRN12–35_3s fragments (GRN-L1, -L2, and -L3) were chemically synthesized on a 0.1 mmole scale using fluorenylmethyloxycarbonyl (Fmoc) solid-phase peptide synthesis. In the truncated analogue GRN-L3, Cys III and Cys V were replaced with alanine residues to prevent disulfide bond formation between these residues and enable the formation of the Cys IV to Cys VI disulfide bond present in Ov-GRN12–35_3s. The folding yields for GRN-L1, GRN-L2, and GRN-L3 were 25%, 33%, and 30% respectively. The sequences of GRN-L1, -L2, and -L3 are shown in Figure 1. In addition, the locations of the loops related to these three fragments are highlighted on the three-dimensional structure of Ov-GRN12–35_3s. Following purification of the peptides with RP-HPLC, the correct masses of the peptides were confirmed using MALDI TOF/TOF and electrospray mass spectrometry (Figure S1). Purified GRN-L1 was very broad despite only displaying one mass. This appears to be related to the additional, minor conformations present, as evidenced in the NMR spectra.
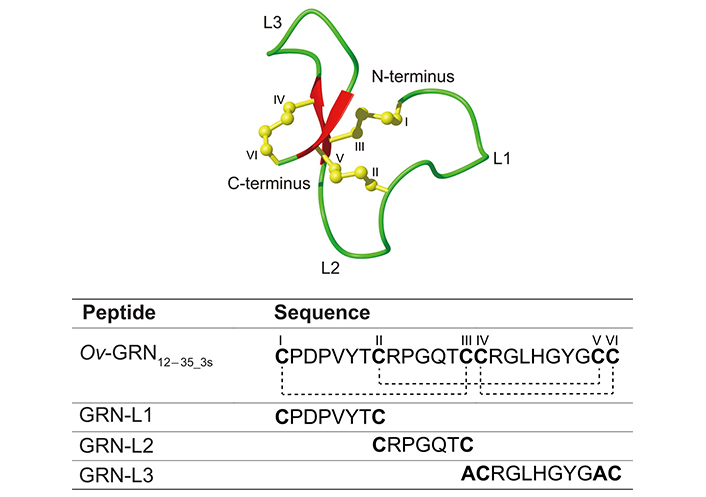
Three-dimensional structure and sequence of Ov-GRN12–35_3s. The sequences of Ov-GRN12–35_3s (PDB ID: 5UJG) and the fragments are given below the structure. The cysteine residues are numbered using Roman numerals (I–VI) and the disulfide bond connections highlighted with dotted lines
Analysis of the one-dimensional (Figure 2) and two-dimensional NMR spectra of the truncated granulin peptides allowed the assignment of the majority of the proton resonances and determination of the secondary shifts (Figure 3A). This analysis indicated that the GRN-L1 and -L3 peptides had no consecutive positive or negative secondary chemical shifts, consistent with the peptides being unstructured in solution. The chemical shifts for the Ov-GRN-1 fragments are given in the Table S1. Although GRN-L2 had consecutive positive secondary shifts for residues 4–7, which are consistent with the presence of β-sheet structure, there was a relatively low number of non-intra-residue or sequential NOEs in the NOESY spectrum, which prevented the determination of a well-defined structure. Three-dimensional structures were calculated with CYANA and confirmed the peptides generally had poorly defined structures, albeit constrained by the single disulfide bond as shown in Figure 3B. Structure statistics are given in Table S2. GRN-L2 appears to have the most well-defined structure as a result of having the smallest ring size.
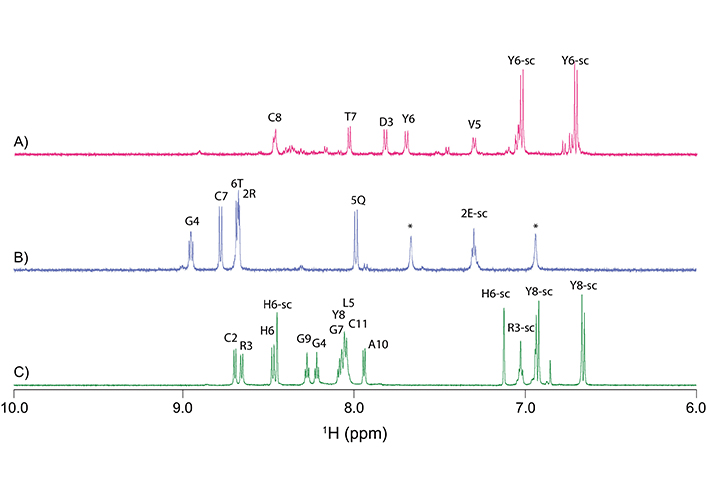
One-dimensional proton NMR spectra of Ov-GRN-1 fragments. A. GRN-L1; B. GRN-L2; and C. GRN-L3. Spectra are shown from 6.5 ppm to 9.5 ppm; this region contains primarily the amide and the aromatic ring protons. Amide protons are labeled with the one-letter code and residue number. Peaks corresponding to side-chain protons are labeled with the one-letter code and residue number and extension -sc. *: NH4+
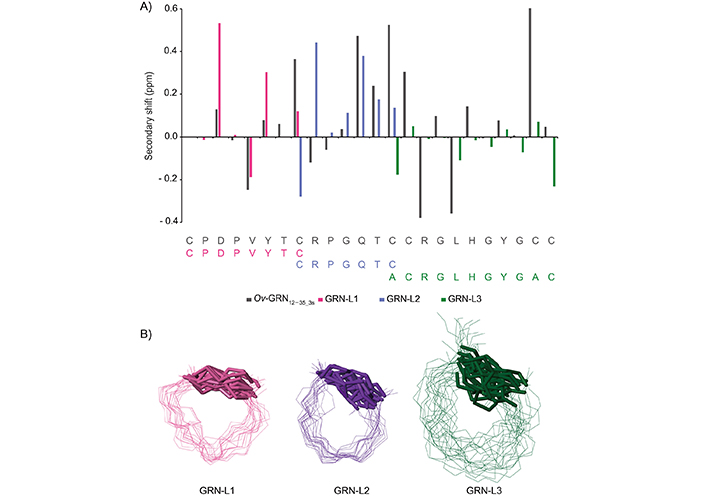
αH Secondary-shifts and three-dimensional representation of Ov-GRN-1 fragments. A. The αH secondary shifts were calculated by subtracting the random coil 1H NMR chemical shifts previously reported by Wishart et al. [21] from the experimental αH chemical shifts. The sequence of the peptides is given at the bottom of the diagram. The Ov-GRN12–35_3s (BMRB code: 30232) shifts are provided as a comparison; B. superposition of the 20 lowest energy structures based on CYANA calculations. The disulfide bonds are shown with thickened lines
The influence of the Ov-GRN12–35_3s fragments on the proliferation of a human normal fibroblast cell line in real time was assessed using xCELLigence technology. GRN-L2 resulted in a 13.6% and 13.7% increase (P < 0.01) in cell growth compared to the negative control peptide when applied to cells at concentrations of 200 nmol/L and 1 μmol/L respectively. A 21-residue tropomyosin fragment was used as a negative control; tropomyosin fragments have previously been shown to have no cell proliferation effects [12, 13, 22]. Importantly, this peptide was produced in the laboratory using the same conditions as the Ov-GRN12-35_3s fragments. GRN-L1 and GRN-L3 peptides tested at concentrations of 200 nmol/L and 1 μmol/L did not display statistically significant cell proliferation compared to the peptide control (Figure 4). GRN-L1 and GRN-L2 were also tested at lower concentrations but the cell proliferation was not statistically significant compared to the control peptide. Data from days 1 to 3 are given in Figure S2. The real-time response of the Ov-GRN-1 fragments at 1,000 nmol/L (Figure S3) highlights the enhanced potency of the GRN-L2 compared to the two other fragments.
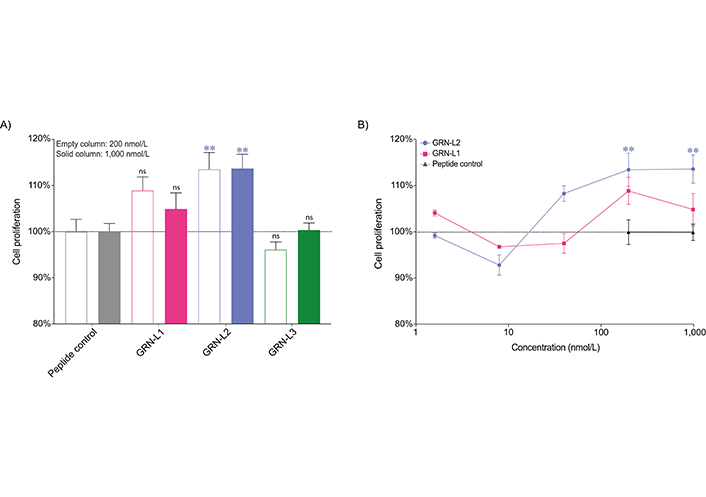
Cell proliferation assay of Ov-GRN-1 fragments. The cell index of the fibroblast cell line, 1BR.3.GN, was measured four days after treatment and the proliferation rates relative to peptide control are plotted as mean ± SEM bars. Data were analyzed by one-way ANOVA against peptide control (ns: not significant, ** P < 0.01). A. Empty columns and solid columns represent concentrations of 200 nmol/L and 1 μmol/L, respectively for the peptide control and Ov-GRN-1 fragments. GRN-L2 displayed significant increases in cell proliferation at these two concentrations, in contrast to GRN-L1 and GRN-L3; B. GRN-L1 and GRN-L2 were tested over a wider range of concentrations but significant enhancements in cell proliferation were only observed for the GRN-L2 peptide at 200 nmol/L and 1 μmol/L. Given the complete lack of activity for GRN-L3 at 200 nmol/L it was not tested at lower concentrations. SEM: standard error of the mean
Truncated forms of Ov-GRN-1 have significant potential as wound-healing agents but the key features for bioactivity are still poorly understood. In the current study a peptide corresponding to the second inter-cysteine loop of Ov-GRN-1 (GRN-L2) was shown to promote cell proliferation of human fibroblasts. This sequence is unique to Ov-GRN-1 in the granulin family based on a BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi), indicating that this loop could be responsible, in part, for the potent wound-healing activity of Ov-GRN-1. The rate of proliferation enhancement is less than the levels observed in previous studies on Ov-GRN-1 and truncated Ov-GRN-1 peptides [12]. However, further studies are required to determine if this rate of proliferation correlates with wound healing in a mouse model, or ultimately with therapeutic potential in humans.
Based on NMR spectroscopy GRN-L2 appears to be unstructured in solution, suggesting that a well-defined structure is not critical for bioactivity. There are now numerous examples of peptides that lack well-defined structures but still display bioactivity. These examples include disulfide-rich peptides, unstructured because of reduction or modification of the cysteine residues, such as human granulin B and the scorpion venom peptide chlorotoxin (CTX). The structural analysis of fully reduced human granulin B indicated that the peptide was intrinsically disordered, but it was able to induce an inflammatory response in SY-SH5Y human neuroblastoma cells by activation of nuclear factor-kappaB in a concentration-dependent manner [23]. Furthermore, CTX contains 36 amino acids and inhibits glioma cell migration [24]. Despite the full-length peptide having a well-defined structure braced by four disulfide bonds, the reduced form of CTX adopts a random coil structure and inhibits cell migration with a similar potency to CTX [25].
Synthetic fragments of CTX with no disulfide bonds have also been studied and a region corresponding to residues 29 to 36 of CTX, which is not structured in solution, inhibited cell migration, albeit at relatively high concentrations [24]. In a parallel study, this region of CTX was used to design blood-brain barrier shuttles, which included a peptide referred to as miniCTX3, a monocyclic lactam-bridge peptidomimetic containing residues 29–32 of CTX. This truncated analogue of CTX was capable of transporting nanoparticles across endothelial cells [26]. Therefore, the identification of a bioactive, inter-cysteine loop of Ov-GRN-1, despite being unstructured in solution, might be useful for further studies in drug design.
CTX: chlorotoxin
NMR: nuclear magnetic resonance
Ov-GRN-1: Opisthorchis viverrini granulin-1
RP-HPLC: reversed-phase high-performance liquid chromatography
The supplementary material for this article is available at: https://www.explorationpub.com/uploads/Article/file/100812_sup_1.pdf.
NLD: Conceptualization. RT, DTW, MJS, PSB and NLD: Methodology, Investigation, Data curation, Formal analysis. RT and NLD: Writing—original draft. NLD, DTW, PSB and AL: Supervision, Resources. All authors discussed the results and commented on the manuscript.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
All datasets for this study are included in the manuscript and the supplementary files.
RT was supported by a James Cook University PhD scholarship. NLD was supported by a
© The Author(s) 2023.
Copyright: © The Author(s) 2023. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Jia Li Guo ... Ji Zhong Zhao
Maria G. Ciulla ... Kamal Kumar
Miguel García-Castro ... Juan Manuel López-Romero
Anton Kolodnitsky ... Vladimir Poroikov
Atri Das ... Shantanabha Das
Galana Siro, Atanas Pipite
Maya G. Pillai, Helen Antony
