Affiliation:
1Núcleo de Biotecnología Curauma, Pontificia Universidad Católica de Valparaíso, Valparaíso 2373223, Chile
Email: constanza.cardenas@pucv.cl
ORCID: https://orcid.org/0000-0002-3673-9320
Affiliation:
2Instituto de Ciencias Aplicadas, Facultad de Ingeniería, Universidad Autónoma de Chile, Santiago 8910060, Chile
ORCID: https://orcid.org/0000-0002-5742-4926
Affiliation:
3Laboratorio de Cultivo de Peces Marinos, Departamento de Acuicultura, Universidad Católica del Norte, Coquimbo 1781421, Chile
4Laboratorio de Fisiología y Genética Marina (FIGEMA), Centro de Estudios Avanzados en Zonas Áridas (CEAZA), Coquimbo 1781421, Chile
ORCID: https://orcid.org/0000-0003-3132-2187
Affiliation:
5Laboratorio de Patógenos Acuícolas, Instituto de Biología, Pontificia Universidad Católica de Valparaíso, Valparaíso 2373223, Chile
ORCID: https://orcid.org/0000-0003-3883-5188
Affiliation:
6Laboratorio de Genética e Inmunología Molecular, Instituto de Biología, Pontificia Universidad Católica de Valparaíso, Valparaíso 2373223, Chile
ORCID: https://orcid.org/0000-0003-0438-8252
Affiliation:
7Department of Organic Chemistry and CIBER‑BBN, Networking Centre on Bioengineering, Biomaterials and Nanomedicine, University of Barcelona, 08028 Barcelona, Spain
8School of Chemistry and Physics, University of KwaZulu-Natal, Durban 4001, South Africa
ORCID: https://orcid.org/0000-0002-8946-0462
Affiliation:
1Núcleo de Biotecnología Curauma, Pontificia Universidad Católica de Valparaíso, Valparaíso 2373223, Chile
Email: fanny.guzman@pucv.cl
ORCID: https://orcid.org/0000-0002-7829-0568
Explor Drug Sci. 2024;2:701–718 DOI: https://doi.org/10.37349/eds.2024.00070
Received: July 09, 2024 Accepted: August 21, 2024 Published: October 15, 2024
Academic Editor: Marcello Iriti, Milan State University, Italy
The article belongs to the special issue Bioactive Peptides: Pioneering Innovations in Latin American Research
According to the International Union of Pure and Applied Chemistry (IUPAC), peptides are small proteins with a size between 2 and 50 amino acids residues. They are ubiquitous across the evolutionary scale, fulfilling a wide variety of functions, from immune system effectors in simple organisms to signaling or neuromodulating agents in high vertebrates. Following nature’s example, peptides have emerged as alternatives in various fields. One particularly relevant area is in drug discovery, offering alternatives to face the emergence of antibiotic-resistant microorganisms. Peptides are also prevalent in other sectors, such as the food industry, where they serve as food additives to enhance nutritional characteristics or aid in food preservation. Moreover, peptides are increasingly being utilized in cosmetics. Additionally, peptides serve as valuable tools in both basic and applied research, facilitating the exploration of specific activity mechanisms and the verification of particular activities, among various other applications. Despite certain limitations and disadvantages compared to other bioactive molecules, peptides remain a focal point of interest in research, as well as in applied and developmental fields, due to their versatility. In this report, we provide an overview of the extensive application landscape of synthetic peptides, presenting examples developed in-house across different areas which include a summary of the methodologies and results obtained.
According to the International Union of Pure and Applied Chemistry (IUPAC), peptides are small proteins with a size between 2 and 50 amino acids residues (https://iupac.qmul.ac.uk/AminoAcid/A1113.html#AA11) and are present in nature performing a wide variety of functions, from effectors of the immune system in low complexity organisms [1, 2] to signaling or neuromodulating agents in mammals [3].
Following nature’s example, peptides have emerged as alternatives in various fields. One particularly relevant area is in drug discovery [4–7], offering alternatives to face the emergence of antibiotic-resistant microorganisms thanks to their multiple ways of action that minimize the possibility of resistance [8–11]. Peptides are also prevalent in other sectors, such as the food industry, where they serve as food additives to enhance nutritional characteristics [12–14] or aid in food preservation [15, 16]. Moreover, they are increasingly being utilized in cosmetics [17–19].
The therapeutic peptide market is estimated at around 45 billion dollars and a growth of 76% is projected by 2032 (https://www.precedenceresearch.com/peptide-therapeutics-market consulted on 08/07/24).
Currently, there are more than 80 peptide-based drugs approved for use, with a growing number of peptides in various phases of approval [20]. Notably, in 2023, the Food and Drug Administration (FDA) approved four peptides for different applications, and with varied structures and diverse administration routes [21], showing the versatility of these molecules.
The projection for the use of peptides and their synthesis aligns with the development of green chemistry for their production, both at the laboratory level and in scaling up their production. Even though the path is just beginning, advances have already been made in this area [22–24].
Additionally, peptides serve as tools in both basic and applied research [25–30], facilitating the exploration of specific activity mechanisms and the verification of activities, among several other applications.
In this report, we provide an overview of the extensive application landscape of synthetic peptides, presenting examples developed in-house across different areas which include a summary of the methodologies and results obtained.
This is one of the best-known areas with multiple applications. The underlying idea is the detection of a protein through recognition by antibodies generated against an epitope of it. This strategy has been used to determine salmonid immune system molecules, among others, and several papers related to this point have been published [31–36].
The procedure followed in our laboratory can be seen in Figure 1.
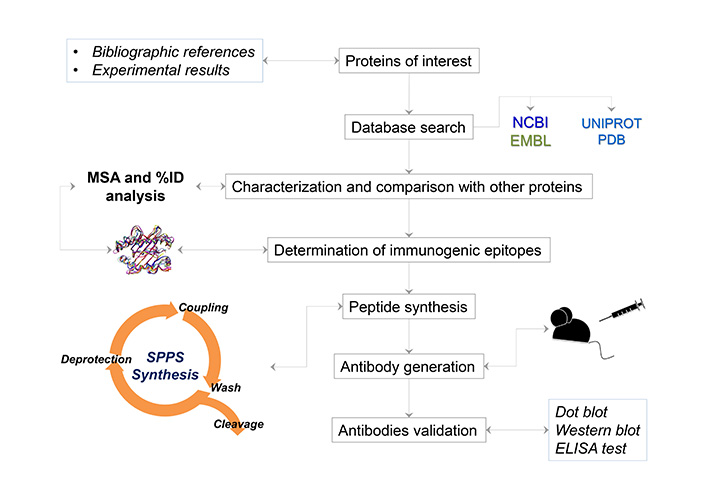
General procedure for obtaining antigenic peptides for the generation of antibodies
The main steps of the methodology are described below.
Once defined the protein target, the amino acid sequence is obtained from databases. Then, conserved sequence motifs should be identified if a cross-reactive antibody is desired, or non-conserved regions if a differential antibody is desired. It is also preferable to have a 3D structure of the protein, which can be obtained through homology modeling, in case of not having an experimental structure. This characterization is complemented by the prediction of antigenic sites; this can be done automatically by accessing different servers, such as:
Emboss antigenic [37]: https://www.bioinformatics.nl/cgi-bin/emboss/antigenic
Predicting Antigenic Peptides: http://imed.med.ucm.es/Tools/antigenic.pl
Antibody Epitope Prediction [38]: http://tools.immuneepitope.org/bcell/
SVMTriP [39]: http://sysbio.unl.edu/SVMTriP/index.php
Additionally, several resources regarding T cell epitope prediction servers are described in the review by Soria-Guerra et al. [40]. The epitope determination is complemented by the analysis of 3D structure, choosing the sequences favoring those found in flexible and exposed regions. 3D structures could be obtained from databases, if they have been experimentally determined, or by homology modeling, which can be obtained through multiple servers such as:
I-TASSER [41]: https://zhanggroup.org/I-TASSER/
RaptorX [42]: http://raptorx6.uchicago.edu/ContactMap/
Phyre2 [43]: http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id=index
SWISS-MODEL [44]: https://swissmodel.expasy.org
The chosen sequences are then chemically synthesized, by fluorenylmethyloxycarbonyl (Fmoc) strategy, using tea-bag protocol and are characterized and purified [23, 45]. At this point it is important to note that there are two strategies that guarantee a good antigenic response:
The generation of the synthetic peptide with cysteine residues in the amino and carboxy terminals, allows polymers of the original sequence to be obtained, guaranteeing better recognition of it in the model in which the antibodies are generated (mice or rabbits) [33]. In this regard, the polymerization of the peptides is performed by oxidation at a low concentration (4 mg/mL), to prevent the formation of high molecular weight polymers, which negatively affect the solubility of the peptide.
The use of the FIS peptide (FISEAIIHVLHSR) a known T helper cell activator in the animal immunization with epitope peptide [46].
These two strategies make it possible to avoid the use of carrier proteins.
Synthetic peptides have been used to generate polyclonal antibodies mainly in mice and rabbit models. As mentioned before, polymers of synthetic peptides are used in the formulation. The inoculum of the peptide with FIS is prepared at a 1 mg/mL concentration and the FIS peptide is added at a 1:1 proportion with respect to the peptide. The first dose is administered with complete Freund’s adjuvant, and then two boosts are given before obtaining ascites fluid or blood for antibody purification. Peptide doses are commonly 50–100 μg per mouse and 300–500 μg for rabbits, while the purification of antibodies is conducted through immunoaffinity chromatography, employing cyanogen bromide-activated Sepharose. Finally, the validation is made by dot blot and ELISA using the epitope-peptide, and western blot using protein extract from a tissue that expresses the protein of interest.
Among the multiple roles that peptides can play their role as immune system modulators stands out. This is especially relevant in the case of the adaptive immunity of mammals and teleost fish. This role has been investigated by us specifically in fish aquaculture species. Regarding this topic, there are peptides, such as hepcidin and cathelicidin, whose primary function in salmonids has been described as antimicrobial. However, when administered in fish or fish leukocyte-type cell lines they upregulate or downregulate pro-inflammatory and/or anti-inflammatory cytokines [47–49]. In the last year, these peptides have been named “host defense peptides” (HDP). Figure 2 shows a summary of the methodology.
Utilizing model homopeptides proves valuable in examining their activity and its relationship with secondary structure. Additionally, they serve as a first step in the development of functional heteropeptides. In previous studies, homopeptides composed of three different amino acids, Pro, Lys, and Arg, were selected to investigate the relationship of secondary structure with antimicrobial and antifreeze activity [50–54]. The amino acids were used to synthesize homopeptides of different chain lengths (7–14) using the Fmoc strategy and tea-bag methodology. Homopeptides were characterized in terms of their secondary structure by using circular dichroism (CD) spectroscopy and tested for their activity. Figure 3 summarizes the methodology used for homopeptides studies.
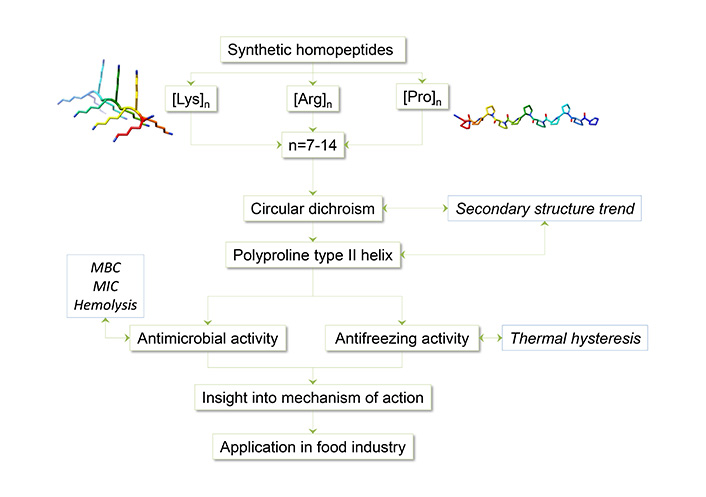
The specific methodology used in this approach is described below.
After the synthesis and cleavage, the lyophilized peptides were dissolved in water at 1 mM concentration and in 30% trifluoroethanol (TFE), and their corresponding CD spectra were measured at different temperatures using a CD spectrometer. Each spectrum was the average of three scan repetitions in continuous scanning mode with 100 nm/min scanning speed with a response time of 2 seconds.
As described in previous work [50–53, 56], the antimicrobial test was performed using Gram+ and Gram– bacterial strains from ATCC or collection to determine their activity. Minimal bactericidal concentration (MBC) or minimal inhibitory concentration (MIC) were determined by the microplate dilution assay. The different bacterial strains were grown to a logarithmic phase at optimum culture media and temperature. Peptides were probed at less than 100 µM concentration and the use of serial dilutions.
To determine the peptides toxicity two kinds of assays were performed: cytotoxicity and hemolysis [32, 52, 53, 56].
The cytotoxicity of the peptides can be determined by exposing cell monolayers, such as Chinook salmon embryonic cells (CHSE-214) to homopeptide concentrations covering values at least ten times higher than MBC. Cell viability was determined by the trypan blue exclusion technique.
The hemolysis assay was performed with blood samples, from healthy human donors or animals, collected in tubes with anticoagulant. A separation of plasma and cells was made by centrifugation at 2,000 × g for 10 min at 4°C. The supernatant plasma was discarded, and the pellet was washed and resuspended in phosphate buffer saline (PBS) to obtain an approximate concentration of 6 × 108 cells/mL. At this point, the peptides to be tested were added in concentrations at least up to 10 times their MBC. A solution of 0.5% triton X-100 in PBS was used as a positive control, while PBS alone was used as a negative control. For the measurement of hemolysis, the samples are centrifuged at 3,000 × g for 5 min. Supernatant samples were taken and their absorbance was measured in a spectrophotometer at 540 nm. The percentage of hemolysis was calculated according to the following equation:
where: As = absorbance of the sample; A100% = absorbance of the positive control; A0% = absorbance of the negative control.
The peptides interacting with bacterial strains can be also detected by confocal laser scanning microscopy using fluorophore-labeled peptide, together with DNA fluorophore probes [51, 57, 58]. Cultures of bacterial strains in mid-log growth phase were incubated with the homopeptides at MBC concentration for 10 to 20 min, and then the fluorescence capture was obtained.
SEM micrograph can be used for the analysis of peptide effects on bacterial membranes [52, 58–61]. Cultures of bacterial strains in mid-log growth phase were incubated with the homopeptides at MIC concentration for 20 min to 1 h. Before microscopic analysis, platinum-palladium powder was sprayed on the samples for better observation of the bacterial membrane structure. By utilizing this technique, the particular residues and structural components that wield the most significant influence in bacterial membrane disruption can be identified.
Different lipid models were used to study the interaction of the peptides with membranes [35, 50–53]. As a model, the use of lipid vesicles formulation with 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), 1,2-dipalmitoyl-sn-glycero-3-phosphorylethanolamine (DPPE), 1,2-dimyristoyl-sn-glycero-3-phospho-(1-rac-glycerol) (DMPG), or with membrane extracts from bacterial strains were prepared by extrusion of frozen and thawed liposome suspensions through polycarbonate filters under nitrogen at 15°C above the lipid transition temperature.
Bacterial membrane extracts can be produced by bacterial pellet sonication, followed by lipid extraction by the Folch method with chloroform/methanol/water mixture. The lipids were obtained by centrifugation at 5,000 × g for 10 min, obtaining the organic phase. Samples were dried under nitrogen and resuspended in MilliQ water at 37°C in a water bath. The lipid suspension was vortexed and sonicated until the turbidity disappeared. Finally, the suspension was used to generate vesicles in the same way as with the model lipids.
A SYTOX Green uptake assay has been widely used to verify the membrane-disrupting ability of the peptides [51, 53, 57–60]. Aliquots of bacterial cell cultures in the exponential growth phase were used to prepare the samples to be read with peptide solutions at different concentrations. Controls include, on the one hand, peptides known to act as cell penetrants peptides and, on the other, membrane-disrupting peptides. The interaction and membrane disruption effects of the peptides were then monitored using a real-time thermocycler or fluorimeter. For penetrating peptides, an increase in fluorescence was not observed, whereas, for membrane-disrupting peptides, an increase in fluorescence was observed.
Hysteresis activity, using a differential scanning calorimeter implemented with an HSS8 high-sensitivity sensor, and a Huber TC45 intracooler chiller, was used to analyze the antifreezing activity of peptides [54]. Peptide solutions at 2, 10, 15, and 25 mM concentrations were used. A total of10 μL of the sample in an aluminum container was cooled down to its freezing point, from 10°C to –25°C at a rate of –3°C/min, then heated at a rate of 3°C/min to –10°C, and then slowly heated at a rate of 1°C/min until partial melting at a recorded temperature (Tho) where the ice coexisted with the peptide solution, which was determined by comparing the cooling curve (where there is recrystallization) and the subcooling curve (where there is no recrystallization). This temperature (Tho) was maintained for 10 min to enhance the interaction between the peptide and the ice crystals. The recrystallization effect of the peptide solution was observed when the temperature was lowered again to –10°C. The next step is to freeze the sample again, cooling it down to –25°C and finally heating it up to 10°C at a rate of 3°C/min to guarantee the return to the initial conditions.
Humanity has always sought its medicines in nature, and peptides are no exception. In addition to those that occur naturally, and have specific functions, there are also “hidden” peptide sequences within the proteins of many organisms that may be bioactive. In this area, peptide synthesis is a valuable tool that allows to focus this search and validate those sequences with potential activity. A summary of the general methodology is presented in Figure 4.
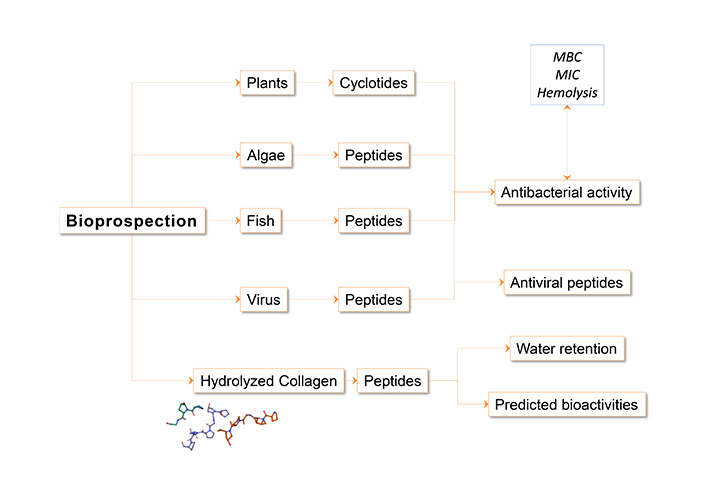
For the selection of the candidate sequences that were synthesized, different methods were used.
Antifungal peptide from hemocytes of Argopecten purpuratus, modified to enhance activity and tested against Fusarium oxysporum, Neurospora crassa, and Saprolegnia sp. [56].
Hepcidin from fish [32, 48, 57, 62]: Hepcidin is a peptide described as antimicrobial in vertebrates which is also found in fish. Taking the sequences available for salmonids, quantitative polymerase chain reaction (qPCR) assays were designed to determine their presence in transcripts from fish stimulated with lipopolysaccharide (LPS); in parallel a polyclonal antibody was generated from the synthetic peptide to detect its presence in tissues. The synthetic peptide was later characterized and tested as bioactive in different contexts.
Cyclotides from Sambucus nigra flower [59, 60]: A peptide extract was obtained from the elder flowers, removing the polyphenols with a polyamide resin. The extract was purified by C18 and the products between 3 and 3.6 kDa were taken to be characterized and tested as antimicrobials. Then mass spectrum analysis allowed the characterization of the cyclotides.
Peptides from the enzymatic digestion of Erythrina edulis (pajuro) flour [63]: Erythrina edulis seed protein concentrate was subjected to hydrolysis with alcalse for 15, 30, 60, and 120 min of incubation. Hydrolysates with antioxidant activity verified by ABTS+ (2,2'-azino-bis(3-etilbenzotiazolin-6-sulfónico)) and peroxyl radical capture Oxygen Radical Absorbance Capacity (ORAC) assays were selected. Active hydrolysates were fractionated by ultrafiltration to select for peptide fragments of molecular weights below 3 kDa. These fractions were purified, and the peptide sequences were determined de novo by mass spectrometry.
Antimicrobial peptides (AMPs) from the microalga Tetraselmis suecica [64]: Cultures of T. suecica were subjected to acid extraction and purified by C18 reverse phase column chromatography using an acetonitrile gradient. The fractions that presented antibacterial activity were characterized by SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) and purified by RP-HPLC (reversed-phase high-performance liquid chromatography), collecting the different fractions where the sequence of the peptides was determined by mass spectrometry.
Cationic peptides as antimicrobials in aquaculture [65, 66]: NK-lysin protein sequences from European sea bass were obtained from UniProt database (https://www.uniprot.org/), and peptide sequences with antimicrobial activity were predicted through the server (http://cabgrid.res.in:8080/amppred/server.php). Additionally, the peptides were characterized in their physicochemical properties and tested for antibacterial and antiviral activity.
Ramosin, AMP identified in Bolitoglossa ramosi (colombian salamander) [61]: From the Bolitoglossa ramosi transcriptome, protein sequences were determined and compared with the APD3 database to select potential antimicrobial AMPs. To obtain candidate peptides, these sequences were analyzed through the ClassAMP predictor server (http://www.bicnirrh.res.in/classamp/predict.php), and the presence of the coding sequences was verified by PCR amplification on cDNA samples of blastemas, skin, and intestine tissues.
Antiviral peptides against viral infections of Atlantic salmon [67]: The infectious pancreatic necrosis virus (IPNV) and infectious salmon anemia virus (ISAV) were the targets for designing interferent peptides. In IPNV, the target process was the formation of the VP2 protein homotrimer, which appears to be the building block in capsid generation, with the peptide interfering with the assembly process of newly formed viruses. In ISAV, the target process was the formation of the RNA-dependent RNA polymerase (RdRp) complex, made up of three different subunits, with the peptides interfering with the formation of a functional RdRp.
Domain of interleukin-8 (IL-8) of salmonids [35, 58]: The protein sequence was analyzed to establish conserved motifs, and their C terminal helical region was selected to be tested as AMP based on information from the literature.
In silico hydrolyzed bovine collagen [68]: Collagen protein sequences from the databases were compared and characterized in terms of their domains and physicochemical properties. These proteins were hydrolyzed in silico with subtilisin, and the obtained peptides were characterized and clustered together with bioactive peptides reported from databases and collected from the literature. Similarity groups were established and candidate peptides with different potential bioactivities were postulated.
Peptide mimicking complement system to eliminate adipocytes [69]: The C3b and CR1 complement system, involved in phagocytosis, was analyzed, and peptides were designed to be coupled with sequences derived from the AQP7 protein as an adipocyte marker, by means of a proline hinge. These hybrid peptides were tested for their ability to bind to adipocytes and promote their phagocytosis.
Bifunctional peptide with adhesive and antimicrobial properties [70]: A bifunctional peptide was designed from a Mytilus edilus protein with adhesive properties, together with an AMP from Bothrops asper. Its use is projected as a coating on implant materials to prevent infections.
After obtaining the sequences, the peptides were chemically synthesized, and their activity verified, with the exceptions of the sequences obtained from elderberry, where activity test was done with the peptide extract as a whole, and in the case of collagen where only the in silico study was carried out.
Regarding the methodology used, there are tests that are transversal, and others that are specific according to the matrix used and the purpose of the test. Below is a summary of the methods that have not been mentioned before.
Additionally, to the methods described in Chemical synthesis, there are some specific processes in the chemical synthesis for particular requirements of peptide sequences, such as cysteine-rich peptides. For example, in the case of hepcidin, which has multiple disulfide bridges, the cysteine with an orthogonal protecting group, Cys-Acm (cysteine-acetoamidomethyl), is used for each pair of cysteines in the sequence that are involved in a disulfide bridge. Four different syntheses were made for each Cys pair (Figure 5) with the goal of directing the disulfide bridge formation. After the synthesis without the final deprotection and prior to the cleavage of the peptide from the resin the Acm group is removed and the peptide is oxidized to generate the specific disulfide bond. Subsequently, the cleavage of the resin is carried out and it is expected that the other disulfide bonds will be generated naturally. With this methodology, it was determined that the most internal bridge between Cys13 and Cys14 was the one that yielded the best result.
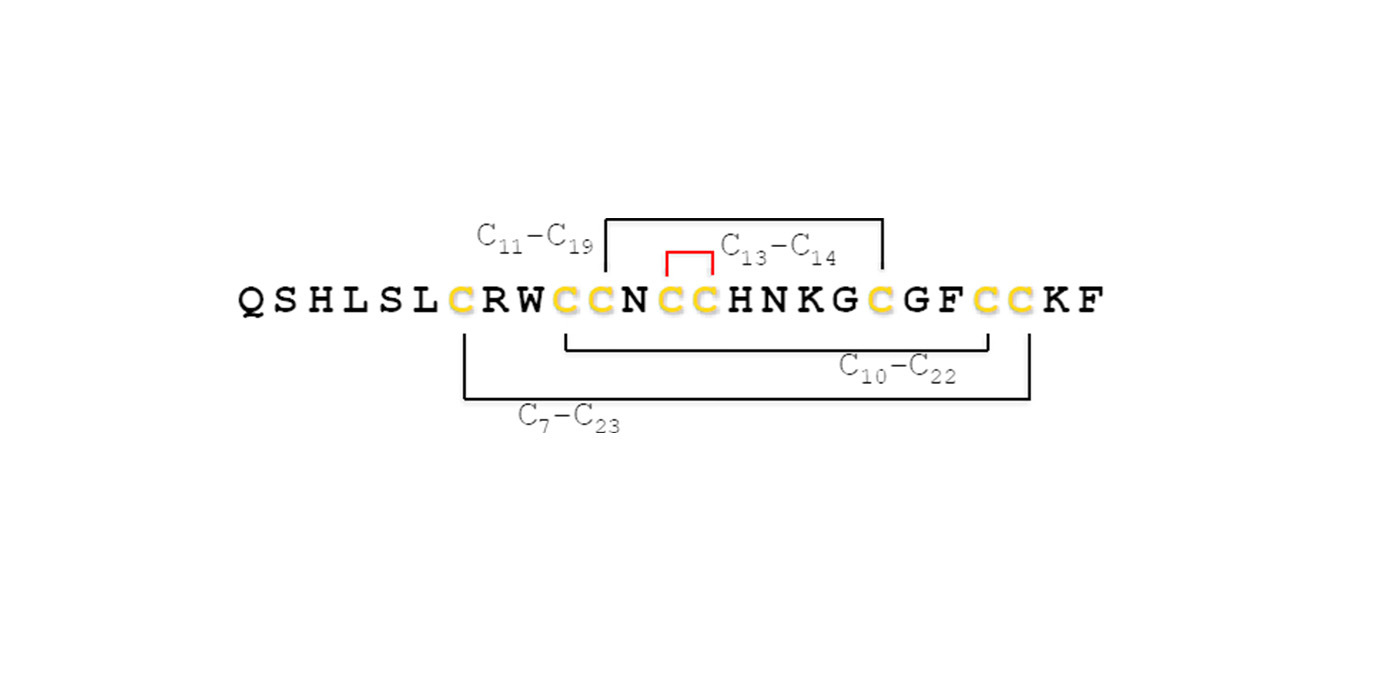
Trout hepcidin sequence with cysteines participating in the disulfide bridges. The initial bond between cysteines 13 and 14 sets the stage for the subsequent formation of the remaining two disulfide bridges
A useful tool to determine important amino acid residues in a peptide sequence is the alanine scan, using this amino acid to mimic the removal of the side chain of each amino acid and thus its specific interactions [71–74]. Tea bag methodology for peptide synthesis is ideal for this approach, being easy to use since there is only one amino acid change per coupling in the set of peptides to be synthesized. These changes allow for verifying the importance of the residue by testing its activity and its influence on the secondary structure when analyzed by CD [52, 53, 64, 75]. One example of the sequence and its analogues by alanine scan is shown in Table 1.
Scan alanine for the R11 homopeptide
| #Peptide | Sequence |
|---|---|
| R11 | RRRRRRRRRRR |
| A1 | |
| A2 | R |
| A3 | RR |
| A4 | RRR |
| A5 | RRRR |
| A6 | RRRRR |
| A7 | RRRRRR |
| A8 | RRRRRRR |
| A9 | RRRRRRRR |
| A10 | RRRRRRRRR |
| A11 | RRRRRRRRRR |
Note. Adapted from Aróstica et al. [53]. © 2022 by the authors
It is possible to use different labels coupled to the peptides, which can be used to track their location by using techniques such as fluorescence, confocal microscopy, or flow cytometry. A summary of the common labels is shown in Table 2. These labels are useful in peptide-specific localization and mechanism of action studies.
Common labels used with peptides
| Label | Structure | Use | References |
|---|---|---|---|
| Rhodamine | 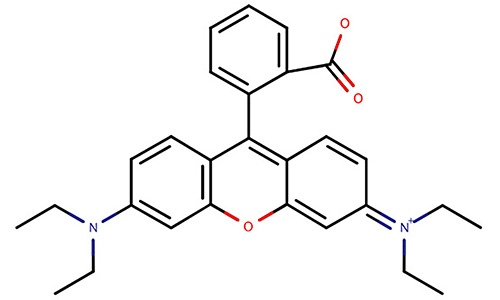 | It can be coupled as an amino acid in the synthesis process and used to track peptide location in microscopy. | [67, 69, 76] |
| Fluorescein | 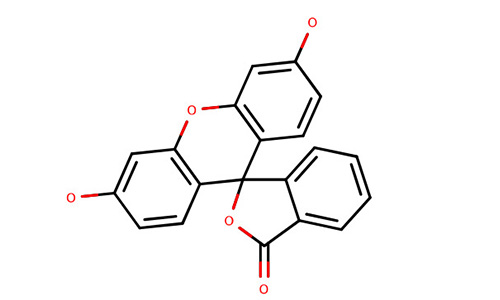 | ||
| Biotin | 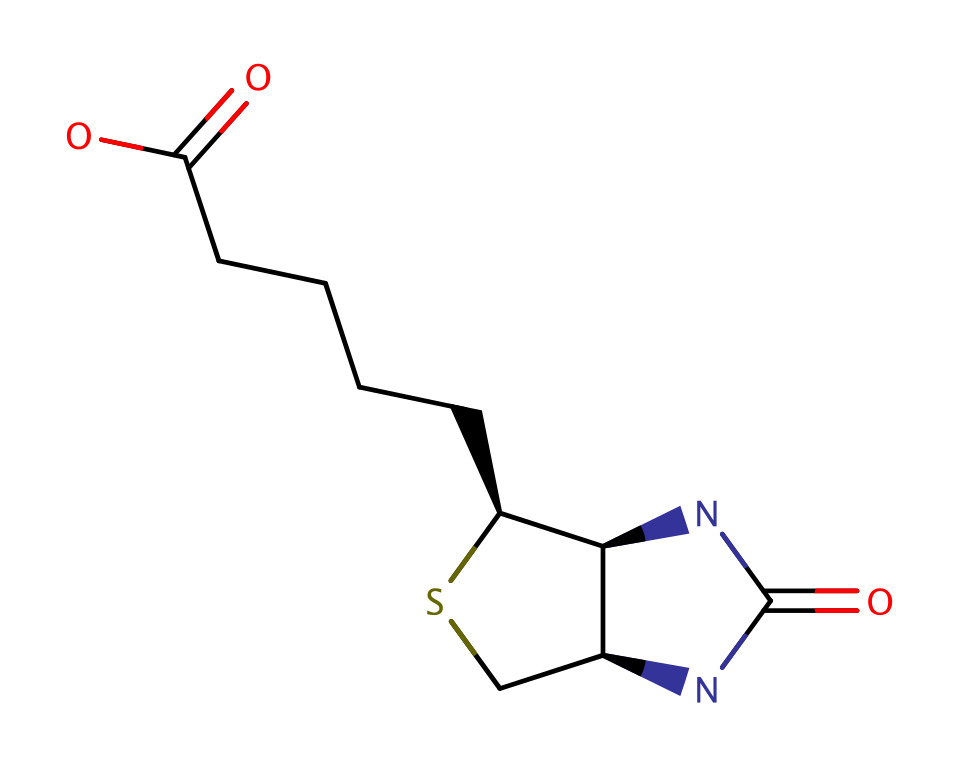 | Coupled as an amino acid in the synthesis process and used to track the peptides by joining them to labeled streptavidin protein in the target cell. |
Compounds structures were drawn with ACD/Chemsketch 2021.2.1 [77]
The synthesis of very long sequences presents a methodological difficulty, since as the peptide chain grows, steric hindrance and agglomeration occur, making it difficult to couple new residues. An option for these sequences is the use of prolines as spacers, taking advantage of the characteristic of this imino acid of changing the direction of the peptide chain, thus reducing the clash between growing chains. It is also possible to use these spacers to generate coupled sequences, or bifunctional peptides. As an example of this a sequence with the capacity to bind to adipocytes and macrophages simultaneously was generated [69].
Finally, there is the possibility of using modified amino acids, to achieve some specific effect. Some examples are summarized in Table 3.
Modified residues or molecules to be used in synthesis for special requirements
| Amino acid | Structure | Use | References |
|---|---|---|---|
| Fmoc-Lys(Fmoc)-OH | 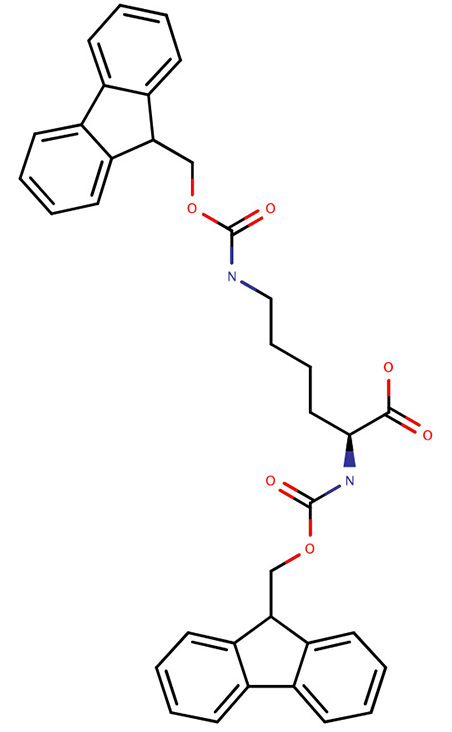 | Synthesis of peptide dimers using the side chain to grow a new peptide chain | [78] |
| Fmoc-Lys(Dde)-OH | 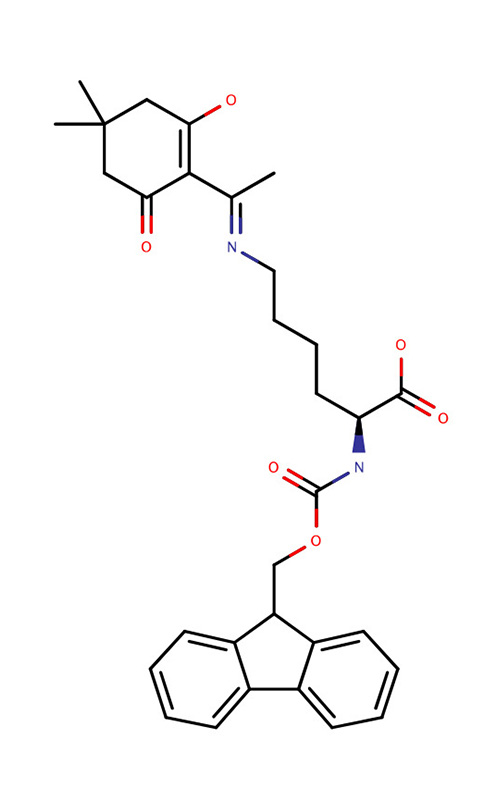 | Used when it is necessary to make bonds with the lysine side chain, for example in the case of branched peptides, template structured peptides, or for bonding with metal groups. | |
| J ligandMalonic or Propanodioic acid | 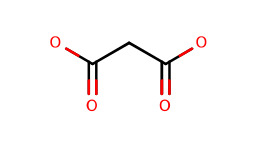 | Used to induce structured peptides in conjunction with Lys(Dde) | |
| Fmoc-DOPA-OH | 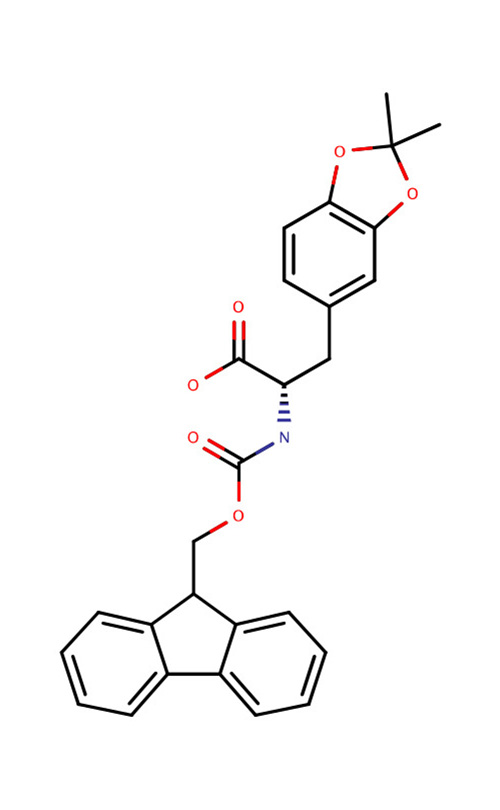 | Catecholic amino acid related to adhesive properties of mussel foot proteins | [70] |
| Deuterated Fmoc-Lys(Boc)-OH | 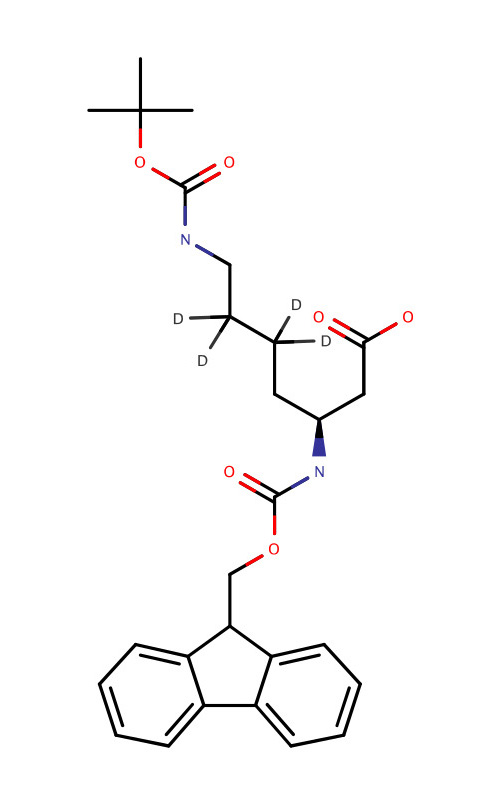 | Track of peptide by nuclear magnetic resonance (NMR) | [79] |
Compounds structures were drawn with ACD/Chemsketch 2021.2.1 [77]. Fmoc: fluorenylmethyloxycarbonyl; Dde: [1-(4,4-dimethyl-2,6-dioxycyclohex-1-ylidine)ethyl]; DOPA: L-3,4-dihydroxyphenylalanine; Boc: tert-butoxycarbonyl
Synthetic peptides have undoubtedly been an invaluable tool, achieving significant advancements thanks to progress in related fields. These include in silico studies to explore diverse applications, structural biology, and new developments in synthesis and delivery methods. As a result, peptides have become a focal point of global interest. An example of a pipeline used for the identification of new active peptides is summarized in Figure 6.
Peptides, as bioactive molecules, have certain advantages and disadvantages compared to other molecules that can perform similar functions, which should be considered in this context. A summary of these characteristics, in comparison to smaller molecules traditionally used as drugs, or larger biomolecules like antibodies or proteins used as therapeutic agents, is presented in Table 4.
| Category | Peptides vs small molecules | Peptides vs biomolecules |
|---|---|---|
| Disadvantages |
|
|
| Advantages |
|
|
Various strategies have been developed to address the limitations of peptides, both in research and production contexts. Numerous studies are continuously conducted based on the specific applications of these molecules. Some of these strategies are summarized in Table 5.
Strategies developed to overcome limitations in the use of peptides
| Limitation | Strategy | References |
|---|---|---|
| Improve stability by preventing proteolysis | Use of modified amino acids such as D amino acids, methylated amino acids, and beta-amino acids among others | [20, 81, 82] |
| Improve stability and activity by preserving secondary structure | Use of hydrogen bond surrogates (HBS), generation of covalent bonds, cyclic peptides | [81–83] |
| Membrane penetration | Conjugation with lipids or biocompatible polymers | [20] |
| Delivery systems to enhance activity and stability | Use of nanotechnologyFunctionalized surfacesNucleic acids systemsHydrogels and scaffolds | [84–86] |
| Cost of synthesis | New strategies towards circular economyProduction as recombinants | [23, 87] |
Peptides are bioactive molecules with a wide range of applications, and thanks to the advancement and integration of numerous research areas, they have become a focal point of global interest. Their versatility in function and the diverse sources from which they can be derived make peptide research a continuously evolving field. In this context, synthetic peptides emerge as a valuable tool, allowing for the precise determination of their functionality, the study of key residues within sequences, and the exploration of potential mechanisms of action. Furthermore, peptides can be synthesized in a short period of time, allowing for changes and testing within reasonable timeframes. This, combined with recent advances in peptide synthesis—moving towards greener and more environmentally friendly processes—along with improved delivery methods to increase their efficacy, ensures that synthetic peptides remain relevant in both basic and applied research. These strategies together can pave the way to improve peptides to achieve specific activities and minimize undesirable effects.
AMPs: antimicrobial peptides
CD: circular dichroism
MBC: minimal bactericidal concentration
PBS: phosphate buffer saline
The authors thank all the students who were part of this journey and who have supported each of the reported publications.
CC: Conceptualization, Data Curation, Writing—original draft, Writing—review & editing, Validation, Visualization, Funding acquisition. PS: Writing—review & editing. CA: Data curation, Writing—original draft, Writing—review & editing, Validation. LM and SM: Writing—review & editing. FA: Writing—review & editing, Validation. FG: Conceptualization, Data curation, Validation, Project administration.
Fernando Albericio, the Editor-in-Chief and a Guest Editor of Exploration of Drug Science, and Fanny Guzmán, a Guest Editor of Exploration of Drug Science, had no involvement in the decision-making or the review process of this manuscript. The other authors declare no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
FG received financial support from ANID Fondecyt [1210056]. CC received funding support form ANID Fondecyt [1240448]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
© The Author(s) 2024.
Copyright: © The Author(s) 2024. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Andrea Barragán-Cárdenas ... Javier García-Castañeda
Natalia Ardila-Chantré ... Javier Eduardo García-Castañeda
Jésica A. Rodríguez ... Silvia A. Camperi
Lucero Ruiz-Mazón ... Mireya de la Garza
Vaezeh Fathi Vavsari, Saeed Balalaie
Vanesa Herlax
Karen Johanna Cárdenas-Martínez ... Javier Eduardo García-Castañeda
Daniela Perdomo-Joven ... Mauricio Urquiza-Martinez
