Affiliation:
1Department of Biology, Eastern New Mexico University (ENMU), Portales, NM 88130, USA
ORCID: https://orcid.org/0009-0001-3398-7003
Affiliation:
1Department of Biology, Eastern New Mexico University (ENMU), Portales, NM 88130, USA
ORCID: https://orcid.org/0009-0000-4061-2050
Affiliation:
1Department of Biology, Eastern New Mexico University (ENMU), Portales, NM 88130, USA
ORCID: https://orcid.org/0009-0001-9682-7314
Affiliation:
1Department of Biology, Eastern New Mexico University (ENMU), Portales, NM 88130, USA
ORCID: https://orcid.org/0000-0003-3017-9112
Affiliation:
2Quality Control Laboratory, Post Harvest Technology, ICAR-Central Institute of Fisheries Education (CIFE), Mumbai 400061, Maharashtra, India
ORCID: https://orcid.org/0000-0001-8600-0025
Affiliation:
2Quality Control Laboratory, Post Harvest Technology, ICAR-Central Institute of Fisheries Education (CIFE), Mumbai 400061, Maharashtra, India
ORCID: https://orcid.org/0009-0004-1441-9539
Affiliation:
2Quality Control Laboratory, Post Harvest Technology, ICAR-Central Institute of Fisheries Education (CIFE), Mumbai 400061, Maharashtra, India
ORCID: https://orcid.org/0000-0003-0223-9069
Affiliation:
1Department of Biology, Eastern New Mexico University (ENMU), Portales, NM 88130, USA
Email: manuel.varela@enmu.edu
ORCID: https://orcid.org/0000-0001-8667-7853
Explor Drug Sci. 2025;3:100897 DOI: https://doi.org/10.37349/eds.2025.100897
Received: December 31, 2024 Accepted: February 27, 2025 Published: March 19, 2025
Academic Editor: Kamal Kumar, Aicuris Anti-infective Cures AG, Max Planck Institute of Molecular Physiology, Germany
The article belongs to the special issue Discovery and development of new antibacterial compounds
Multiple drug-resistant Staphylococcus aureus bacterial pathogens are causative agents of serious infectious disease and are responsible for significant morbidity and mortality rates. Of particular concern in the public health domain are strains of methicillin-resistant S. aureus (MRSA), a member of the Enterococcus faecium, S. aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter spp., Escherichia coli (ESKAPEE) group of bacterial pathogens, many of which are recalcitrant to effective chemotherapy in the clinical setting due to their resistance to multiple antimicrobial agents. An important mechanism that confers multi-drug resistance in MRSA involves the active efflux of structurally different antimicrobial agents by members of the major facilitator superfamily (MFS) of proteins. The multidrug efflux pumps of the MFS share similar amino acid sequences, protein structures, and a common evolutionary origin. As such, the multidrug efflux pumps of the MFS are thought to operate by a similar solute transport mechanism and, thus, represent suitable targets for modulating their transport activities. This review article addresses MRSA as a serious pathogen, the mechanisms of antimicrobial resistance, and the functional and structural roles of the multidrug efflux pumps of the MFS in conferring pathogenicity.
Staphylococcus aureus microorganisms are Gram-positive facultative anaerobic bacteria measuring 0.5–1.0 μm, often visualized as clusters under the microscope and classified as coccus or spherically shaped prokaryotes [1, 2]. This microorganism is a ubiquitous part of the normal flora growing asymptomatically on human skin and mucous membranes, with carriage rates ranging from 20% up to 40% of the human population [2–4]. It also colonizes the anterior nares, vagina, and gastrointestinal tract of nearly one-third of the population without symptoms [5], where the bacterium can, however, be a frequent cause of food poisoning [6, 7].
S. aureus is generally highly resistant to physical and chemical agents, including desiccation, which allows them to adapt to various environmental conditions [7, 8]. In human hosts, S. aureus produces multiple types of enzymes, including α-hemolysins, which contribute to its toxicity, especially in clinical strains [7, 8]. An important distinction among staphylococci is their ability to clot plasma due to the active production of coagulase [9]. Coagulase-positive staphylococci produce coagulase, while coagulation-negative staphylococci do not, and coagulase production is used by the pathogen for virulence and by investigators for identification and classification. The coagulase-positive group includes specific S. aureus strains, plus Staphylococcus intermedius and related species Staphylococcus delphini, and Staphylococcus pseudintermedius [7, 10]. Certain strains of S. aureus, however, are opportunistic infectious pathogens responsible for minor skin infections and severe life-threatening systemic diseases such as pneumonia, endocarditis, and sepsis [7]. S. aureus is a principal causative agent of community- and healthcare-associated bacteremia, infective endocarditis, skin and soft tissue infections, and food poisoning, and is a common pathogen of implantable prosthetic devices, including joint replacements, heart valves, and central nervous system shunts [3, 11]. Strong evidence supports the notion that antimicrobial resistance, aging populations, increased numbers of individuals with immunosuppression, and high prevalence of invasive diseases, such as iatrogenic infections associated with catheter use or surgical implants, account for the global rise in S. aureus-related clinical diseases [4, 12, 13]. As such, S. aureus is a member of the ESKAPEE group of bacterial pathogens, a term first proposed by Rice et al. [14] to encompass Enterococcus faecium, S. aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp., and more recently modified to include Escherichia coli [15], all microorganisms of which are considered serious pathogens of widespread public health concern. It was reported that more than 30 individual bacterial pathogens accounted for more than 7 million deaths in 2019 [16]. About half of these deaths were caused by five pathogens: S. aureus, E. coli, Streptococcus pneumoniae, K. pneumoniae, and P. aeruginosa [17]. Strikingly, S. aureus infections were responsible for over a million deaths globally in 2019, making this microorganism one of the most common causes of bacterial-related deaths in 135 countries and was linked to the most deaths of individuals over the age of 15 years [16, 17]. Most of these deaths are associated with infections of the bloodstream, the lower respiratory tract, peritoneal linings, and intra-abdominal tissue [16, 17]. The burden of these infections is particularly excessive in low- and middle-income countries, where access to effective antimicrobials and healthcare systems is often limited [2, 16, 17].
Certain populations are at a higher risk of S. aureus infections, including individuals with weakened immune systems, those with chronic illnesses, and people who are in close contact with healthcare facilities [4, 10, 18]. Consumption of contaminated food, especially when prepared under unsanitary conditions, can lead to S. aureus food poisoning. Additionally, athletes, military personnel, and people living in crowded or unsanitary conditions are also at an increased risk of infection [19]. Although less common, S. aureus can be disseminated through respiratory droplets, particularly in healthcare settings. Poor hygiene practices and inadequate cleaning of medical equipment and facilities can also contribute to the spread of S. aureus infection. Individuals associated with underlying medical conditions such as diabetes, immunosuppression, and chronic skin diseases are at increased risk of S. aureus infections [19, 20].
Within a decade of the introduction of penicillin, staphylococci resistant to the antimicrobial were discovered, mediated by the production of penicillin-degrading penicillinase, which prompted the development of semi-synthetic, penicillinase-resistant β-lactam antimicrobials such as methicillin, oxacillin, cloxacillin and dicloxacillin in the late 1950s [21]. However, the first methicillin-resistant S. aureus (MRSA) strain was reported from a UK hospital in 1961, followed by Australia, the USA and Denmark [22, 23]. The methicilin-resistance phenotype has been attributed to the acquisition of the mecA gene located on a mobile genetic element, the staphylococcal cassette chromosome mec (SCCmec) [23, 24]. The incidence of MRSA started increasing in industrialized countries in the 1980s, with the emergence of new lineages of MRSA, and the latter part of the 1990s witnessed the emergence of community-associated (CA)-MRSA [10, 25]. Healthcare-associated or hospital-acquired (HA)-MRSA and CA-MRSA are distinguished by SCCmec typing, antibiotic resistance profiles, distribution of virulence genes, and other determinants [25]. The third group, livestock-associated (LA)-MRSA, is genetically distinct from HA and CA groups and is predominantly associated with farm animals. Farm workers are vulnerable to infections by LA-MRSA. However, with the mixing of isolates from different sources and genetic exchanges, the distinctions between these three groups of MRSA have blurred, with clonal types belonging to different MRSA groups being reported from all three environments [25].
The prevalence of MRSA varies geographically and by population group. Depending on the particular region and population, however, the frequency of MRSA associated with human infections can vary widely from less than 1% to over 74% [8, 18, 26]. In healthcare settings, MRSA prevalence can exceed 50%, particularly in intensive care units and among patients with prolonged hospital stays [11]. In the developed world, it is the most common cause of infections following surgery or hospitalization [27]. It is also a prominent cause of wound infections and toxic shock syndrome. Additionally, MRSA is frequently implicated in pediatric skin and soft tissue infections [7].
Concerns were raised by the advent of MRSA, especially in community settings throughout the 1990s. This emergence led to increased MRSA surveillance and the introduction of measures to stop MRSA transmission in such settings [28, 29]. MRSA was primarily spread in clinical or healthcare settings from its discovery in 1961 until the 1980s. LA-MRSA was first discovered in domestic animals in the early 2000s, and its distribution was aided by the food and production systems that house livestock [30].
According to Maguire et al. [31], 3,315 cases of S. aureus illnesses were reported at the Royal Darwin Hospital in Australia between January 1991 and July 1995. In the UK, 13,912 cases of S. aureus bacteremia were reported between April 2022 and March 2023, according to the UK Health Security Agency (HSA), with MRSA accounting for 787 (5.7%) of these cases [32].
MRSA continues to be a major cause of morbidity and mortality in North America, as well. An estimated 119,247 cases of S. aureus septicemia were reported in the United States in 2017, which led to an estimated 19,832 MRSA-related deaths [33, 34]. Kourtis et al. [34] also found that although hospital-onset MRSA bloodstream infections have become much less common as a result of enhanced infection control procedures, CA-MRSA infections are still a problem, particularly for vulnerable groups such as those who suffer from opioid use disorders.
Patients with either prior MRSA history, those with indwelling catheters, hospitalization, and history of surgery, and those with MRSA infection 48 hours after admission are classified as HA-MRSA. Further, patients without prior exposure to a hospital environment in the last six months, without S. aureus infection, have not been on any antimicrobial in the past month, and have no vascular catheter as at the time of infection but have an MRSA isolate with two days of admission are classified as CA-MRSA [35, 36]. It has been reported that healthcare settings account for 85% of all severe instances of the infection, with the remaining 15% of reported infections thought to have been acquired in the community. Individuals who are elderly with poor immunity, experience prolonged hospitalization, and possess other risk factors have higher chances of acquiring HA-MRSA. In contrast, CA-MRSA strains are predominant among young and healthy individuals without risk factors [37].
CA-MRSA spread in the 1980s and was confined to closed communities. In the 1990s, however, CA-MRSA emerged in the healthy population. In the 2000s, a CA-MRSA strain appeared in the US and was denoted USA300-SCCmec IV. Preeja et al. [38] obtained a few hospital-associated MRSA strains in India; it was reported that 31.4% (16/51) were designated SCCmec type IV, and 25.5% (13/51) were SCCmec type V. It has been suggested that hospital environments have become contaminated by CA-MRSA and that community members are the primary source of MRSA circulation, with relatively lower incidence rates of HA-MRSA [39]. The clonal lineages of MRSA are defined by multilocus sequence typing (MLST), and the strains belonging to the same sequence types (STs) belong to the same clonal lineage [40]. Supporting evidence was provided after employing whole-genome sequencing by Chen et al. [41], who discovered that ST59 (the primary clone of CA-MRSA) had supplanted ST239 (HA-MRSA) as the epidemic type of MRSA in China. As a result, CA-MRSA steadily overtakes HA-MRSA as the predominant MRSA infection type.
S. aureus is a highly prevalent bacterium worldwide, with both community-acquired and healthcare-associated strains causing infections. CA-MRSA has also become increasingly common, with notable outbreaks occurring in schools, military settings, and athletes [42, 43]. CA-MRSA spreads more readily through skin-to-skin contact and sharing of personal items like towels or razors [11, 44]. HA-MRSA often spreads through contact with infected wounds or invasive devices such as catheters. Several studies have shown that the prevalence of nasal carriage of S. aureus, a major reservoir for infection, can range from 20% to 30% in the general population, with higher rates in certain high-risk groups such as healthcare workers, patients in healthcare facilities, and individuals with chronic skin condition [4, 5, 12, 13]. The incidence of S. aureus infections has been on the rise, particularly with the emergence of antimicrobial-resistant strains such as MRSA [12, 13]. MRSA has become a major public health concern as a causative agent of severe and difficult-to-treat infections. The pathogenic success of S. aureus is due to an array of secreted and surface structures that enable colonization, immune evasion, and immune suppression [8, 31]. A variety of virulence factors influence the severity of these infections and have been reviewed elsewhere [12, 13].
The emergence of multidrug-resistant (MDR) strains, particularly MRSA, poses a significant public health challenge globally. The rise of MDR S. aureus strains [20], which are resistant to multiple antibiotic classes, including macrolides, aminoglycosides, and fluoroquinolones, makes treatment options significantly complicated, contributing to the enhanced rates of morbidity and mortality [45–47]. As discussed above, resistance mechanisms include the attainment of resistance genes through horizontal gene transfer and mutations in target sites. MRSA, for instance, carries the mecA gene, which encodes a penicillin-binding protein 2a (PBP2a) with a low affinity for β-lactam antibiotics, rendering these agents ineffective [12, 13]. Infections caused by antimicrobial-resistant MRSA require more complex and costly treatment regimens, often involving the use of last-resort antimicrobial agents like vancomycin or linezolid [10, 18]. The ST80 clone in Europe, plus strains USA300 and USA400 of S. aureus in North America, and numerous other strains such as ST59 in Asia and ST30 and ST8 in South America are just a few of the notable examples of the diverse range of CA-MRSA strains [12, 20]. In particular, S. aureus belongs to the ST45 lineage, among others, and is widely distributed across the globe. These strains exhibit resistance to multiple antimicrobial agents, including methicillin, and are notably prevalent in infections related to hospitals and community disease [4].
It has been demonstrated that clonal complex 45 (CC45), which is a cluster of S. aureus isolates defined by MLST, like ST45 and others, branches out close to the origin of the S. aureus population [48]. CC45 is distinguished by its heterogeneity, encompassing both methicillin-susceptible and MRSA strains, community-acquired and hospital-associated strains, as well as clinical and commensal clones. Additionally, strains of CC45 are primarily found in North America, Australia, and Europe despite reports of being less common in Asia, Africa, and South America [49–51]. There are multiple clinically significant MRSA clones in CC45. ST45-MRSA-II, also known as MRSA-USA600, is a common strain in North America and has been documented in Australia and Hong Kong [52]. USA600 is a leading cause of bloodstream infections and endocarditis in North America and is associated with high mortality rates [4, 34].
These strains have specific resistance profiles and genetic traits. One particular characteristic of S. aureus that makes treatment and removal more difficult is host-associated biofilms [1, 16, 53]. Resistance or intolerance to methicillin is correlated with a constitutive strong biofilm-forming trait, which has been proposed as one reason for the increased pathogenicity observed among MRSA [1, 53]. Its contribution to wound contamination, persistence, and biofilm formation at disease sites makes S. aureus a particularly challenging pathogen to treat when infected and persistent biofilms are involved [27]. The MRSA epidemic has prompted an enormous effort to develop new vaccines and antimicrobial agents directed against S. aureus.
To rapidly identify and contain outbreaks, healthcare facilities must maintain constant surveillance and report certain S. aureus strains, including MRSA, to the Centers for Disease Control and Prevention (CDC), which entails monitoring infection rates among staff members and patients as well as screening high-risk patients [54, 55]. Guidelines for prescribing antimicrobial agents and encouraging the prudent use of antimicrobials in medical facilities are part of the measures to reduce the rate at which resistance develops [56]. Decolonization of S. aureus carriers, particularly MRSA, can be done in some situations to lower the risk of infection and transmission. Topical antimicrobials such as mupirocin and antiseptic washes (chlorhexidine) can be used to reduce transmission rates [55]. Infection risk can be mitigated by preventive measures, such as frequent hand washing, covering and cleaning wounds, and refraining from sharing personal objects. Infection control procedures are also crucial in halting the spread of S. aureus [57, 58].
Furthermore, surveillance and pattern recognition of antibiotic resistance are essential for managing MRSA infections effectively. Strict infection control measures must be carried out in healthcare facilities to stop S. aureus from spreading between patients and healthcare personnel. Public health campaigns that emphasize good personal hygiene and responsible use of antibiotics are crucial in community settings. Continued research investigations and surveillance are necessary to track patterns in S. aureus infections and to guide public health initiatives meant to stop and restrict the spread of this serious human pathogen [32, 54].
As a result of genetic modifications and transfer, microorganisms have developed a variety of resistance mechanisms to antimicrobial agents (Figure 1). These mechanisms vary from alteration of antimicrobial targets, antimicrobial inactivation by enzymes, protection of antimicrobial targets, reduced drug entry through the membrane to the cytoplasm, and active efflux of antimicrobial agents from the cytoplasm of bacterial cells [45, 47, 59].
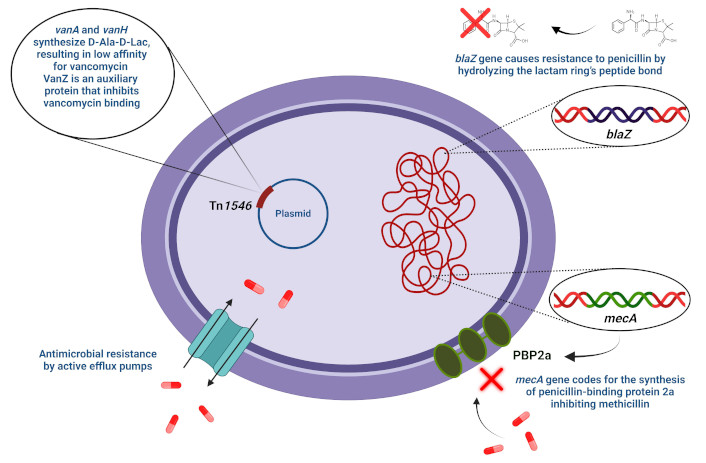
Staphylococcal mechanisms of antimicrobial resistance. D-Ala-D-Lac: D-alanyl-D-lactate; PBP2a: penicillin-binding protein 2a. The red × (top) on the penicillin molecule indicates a form made inactive by the enzymatic action of β-lactamase. The red × (near PBP2a, bottom right) indicates that the mecA-encoded form of PBP2a is unable to bind methicillin, making the cell resistant to the antimicrobial agent
Enzyme degradation is a primary resistance mechanism of S. aureus against penicillin. β-lactamase enzymes encoded by the blaZ gene cause resistance to penicillin by hydrolyzing the lactam ring’s peptide bond, decreasing penicillin’s efficacy in binding to the PBPs [60–62]. The genetic determinant, blaZ, which encodes β-lactamase, is transferable and is encoded on the bacterial genome or carried by large plasmids [63]. The regulatory genes blaR1 and blaI can break to induce the signaling pathway responsible for β-lactamase synthesis. The antirepressor BlaR1 and the repressor BlaI are regulatory proteins that influence the gene expression of β-lactamase [64–66].
The mecA gene, chromosomally localized, is necessary for methicillin resistance. Weighing 78 kDa, the synthesis of PBP2a is the functional end-product of mecA. The mecA gene encodes an altered PBP that is less sensitive to methicillin, producing resistance [67–69]. The PBP enzyme catalyzes the transpeptidation reaction needed for cross-linking peptidoglycan chains. This cell wall construction machinery is believed to have evolved from serine proteases as they share similar activity [61, 62]. By attaching to and inactivating the PBPs in the bacterial cell wall, methicillin impedes the transpeptidation stage of the peptidoglycan formation. The SCCmec, housing the methicillin resistance determinant mecA, is required by S. aureus to confer resistance to this antimicrobial agent.
Vancomycin has historically been recognized as the ultimate protection against infections caused by Gram-positive cocci [70]. A primary mechanism of vancomycin antimicrobial action is the specific binding of the antimicrobial to the bacterial cell wall through small peptides known as peptidoglycan precursors, which have D-alanyl-D-alanine dipeptides at the pentapeptide terminus [71]. This dipeptide-binding by vancomycin prevents the peptidoglycans in the bacterial cell wall from elongating and cross-linking, which represses the synthesis of the cell wall and eventually results in bacterial death [72, 73]. Hence, vancomycin is an inhibitor of bacterial cell wall synthesis [46].
Transposon Tn1546 encodes a vancomycin resistance gene on a genetic locus denoted as the vanA operon [74]. The operon can be found on conjugate plasmids harboring genetic elements denoted as vanA, vanH, vanX, vanS, vanR, vanY, and vanZ [46, 75]. Two methods exist for the horizontal transfer of the vanA operon: the translocation of Tn1546 transposon between distinct plasmids and the transmission of Tn1546 in variant plasmids among strains with different clonal backgrounds [76]. The D-alanyl-D-lactate (D-Ala-D-Lac) ester bond formation is catalyzed by VanA, a ligase, and VanH, a dehydrogenase, which reduces pyruvate to create D-Lac. VanA and VanH synthesize D-Ala-D-Lac, a depsipeptide, which results in a low affinity for glycopeptide antibiotics. VanZ is an auxiliary protein that inhibits the binding of glycopeptide antibiotics to bacterial cell surfaces, hence protecting bacteria from them [76–79]. The relative effectiveness of vancomycin against S. aureus is grouped into three main categories, namely: vancomycin-resistant S. aureus (VRSA) [80], vancomycin-intermediate S. aureus (VISA) [81], and heterologous-VRSA (hetero-VRSA) [82]. Clinical isolates of S. aureus were considered VRSA if they had a minimal inhibitory concentration (MIC) of 32 mg/L or more to vancomycin [80]; VISA bacteria had an MIC to vancomycin 8–16 mg/L [81]. Hetero-VRSA refers to the primary culture of S. aureus isolated from clinical specimens in which sub-populations have varying degrees of susceptibility to vancomycin [83]. However, these types of S. aureus strains are more complicated if MIC is considered as the basis for categorization. The classification scheme takes into consideration the kind of resistance machinery harbored by S. aureus isolates [81, 84, 85].
Biofilms are groups of microbial cells that can attach to surfaces of biological materials for quick adaptation [86]. Due to its components, biofilms decrease the permeability of antimicrobial agents. Microorganisms in biofilms slow down their growth rates to avoid interaction with and the effects of antimicrobials. However, specific resistance genes can be present in biofilms, all of which contribute to the complexity of the mechanism of biofilm-mediated drug resistance. Also, antimicrobial agents have difficulty entering and spreading through biofilms because of their barrier properties, drastically lowering antimicrobial absorption [87–90]. Lastly, large numbers of persistent cells in biofilms allow bacteria to block targets on their surface and maintain a low metabolic state, shielding them from the effects of antimicrobials and fostering MDR.
MDR efflux pumps are bacterial transport systems that can actively extrude multiple classes of antimicrobial chemicals or are selective, extruding only one or a class of antimicrobial agents [91]. Although antimicrobial agents can help reduce the effectiveness of bacterial infection, as a result of the efflux of antimicrobials, MDR bacteria can acquire resistance to chemotherapeutic agents. Antimicrobial efflux pumps of MRSA are grouped into five principal protein superfamilies of related transporters: the major facilitator superfamily (MFS), the small multidrug resistance (SMR) family, the multidrug and toxin extrusion (MATE) family, the adenosine triphosphate (ATP)-binding cassette (ABC) superfamily, and the resistance nodulation division (RND) superfamily [30, 92–96]. Depending on how drug transporters derive their energy, they are grouped into primary and secondary active transporters. While the primary transporters rely on the energy of ATP hydrolysis, the secondary active transporters rely on concentration variations formed from ions, such as protons or sodium, across the membrane. Importantly, secondary active transporters are mostly antimicrobial drug targets because of their recognition sites and substrate selectivity profiles [97, 98].
One of the largest protein families of membrane transporters is the MFS, which is involved in both the efflux of antimicrobial agents and the substantial increase in bacterial MDR [99]. Members of the MFS that are antimicrobial efflux pumps include NorA, NorB, NorC, LmrS, EmrD-3, TetA(K), TetA(L), MdeA, SdrM, QacA, and QacB [100]. Many of the MFS transporters typically consist of 12 transmembrane helices in which the N- and C-termini reside on the cytoplasmic side of the bacterial membrane (Figure 2). Such efflux pumps of the MFS with 12-transmembrane segment (12-TMS) in S. aureus include NorA [101, 102] and NorB [103].
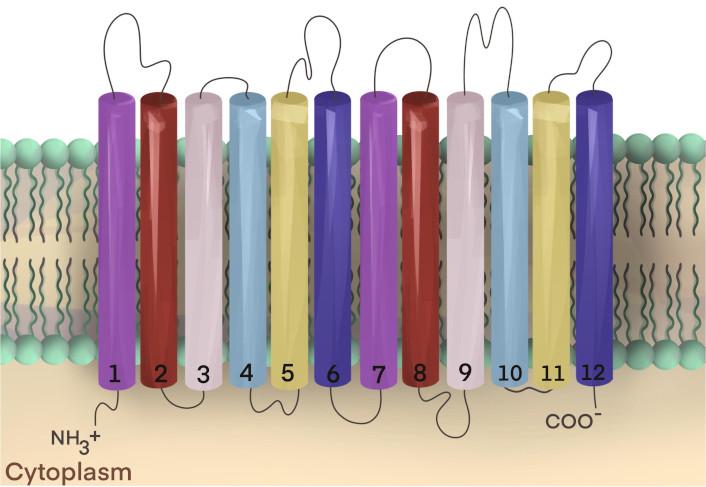
Secondary structure of 12-transmembrane segment (12-TMS) transporters. Solute transporters of the major facilitator superfamily (MFS) share a common secondary structure in which the N-terminal and C-terminal reside in the cytoplasm of the cell and transverse the biological membrane as 12 hydrophobic α-helical domains called TMSs that are separated by interhelical loops [104]. Transport proteins of the MFS from Staphylococcus aureus with 12-TMS include NorA and NorB [96]. This figure is adapted from Lekshmi et al. [99] (CC BY 4.0)
This 12-TMS secondary structure pattern is evident in both passive and active transporters of the MFS [105]. These protein members of the superfamily share similarities in amino acid sequences and include symporters of non-antimicrobial solutes, as well as antiport-based antimicrobial efflux pumps [106]. One symporter in particular, the lactose permease, LacY, has served as a model system for the investigation of solute transport across the membrane [107]. Efflux pumps (especially those belonging to the MFS) are essential to S. aureus MDR [108].
Transporters of the MFS are also thought to harbor 14-TMS across the bacterial plasma membrane, including antimicrobial efflux pumps of S. aureus (Figure 3) [109]. The transporters of the MFS function via a proton or sodium (ion) motive force, using an electrochemical gradient of ions across the cell membrane to export antimicrobial substances out of the bacteria cell, thus diluting the internal cytoplasmic concentration in the cytoplasm where the antimicrobial targets chiefly reside [110, 111]. These solute pump systems can expel a wide range of antimicrobial agents, including methicillin, tetracyclines, chloramphenicol, and fluoroquinolones [112].
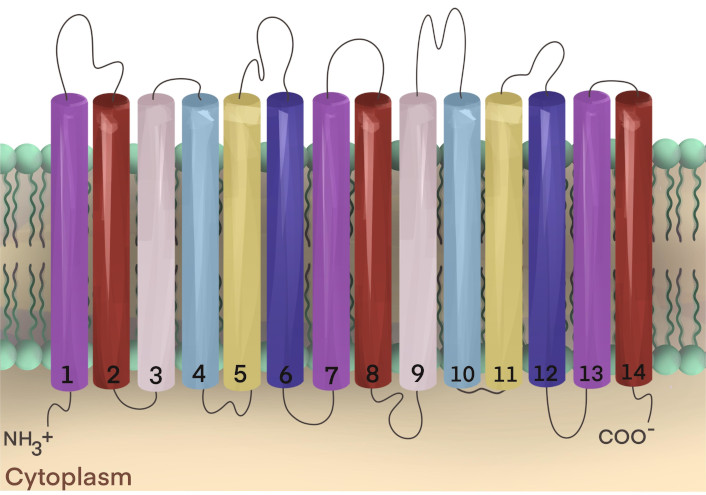
Secondary structure of 14-transmembrane segment (14-TMS) transporters. Transporters of the major facilitator superfamily (MFS) that harbor 14-TMS in Staphylococcus aureus have their N- and C-termini located inside the cell, on the cytoplasmic side of the membrane. The TMSs are connected by inter-helical loops that reside between the putative transmembrane α-helices. Characterized MFS transporters from S. aureus include Tet38, QacA, QacB, MdeA, SdrM, LmrS, TetA(K), TetA(L), and NorC [96]. The two-dimensional image is adapted from Lekshmi et al. [99] (CC BY 4.0)
Proteins that are members of the MFS share similarities in amino acid sequences, as evidenced by the presence of highly conserved sequence motifs. Furthermore, these transporters share similarities in molecular configurations from a structural perspective, suggesting that these proteins have a common ancestral or evolutionary origin [113]. For MFS transporters with 12-TMS, the N-terminus bundles consist of the first six transmembrane α-helices, and the C-terminus domains have α-helices 7 through 12 [113], while for the MFS proteins that possess 14-TMS, the N- and C-termini consist of α-helices 1–7 and 8–14, respectively. These structural domains have been referred to as the MFS fold [114].
Whether the various MFS transporters harbor 12- or 14-TMS, overexpression of these pumps is often linked to antimicrobial resistance in pathogenic bacteria like S. aureus, E. coli, and P. aeruginosa [115]. The NorA multidrug efflux pump is one of the most studied MFS pumps in S. aureus. NorA is known to export fluoroquinolones, biocides, and disinfectants [101, 102, 116]. MRSA infections result in higher morbidity, mortality, and healthcare costs, and the presence of MFS efflux pumps not only contributes to treatment failure but also complicates the development of new antimicrobial agents [117].
In S. aureus, NorA is among the first antimicrobial efflux pumps to be studied from molecular and physiological perspectives [116]. The bacterial genome harbors the norA drug resistance determinant [101]. NorA confers resistance to hydrophilic fluoroquinolones (ciprofloxacin, norfloxacin). NorB and NorC confer resistance to hydrophobic fluoroquinolones (moxifloxacin and sparfloxacin) [118].
The efflux protein, TetA, is encoded by the tet gene. Mostly found on tiny transmissible plasmids, the tetA resistance determinants are occasionally integrated into staphylococci’s chromosomes, promoting the development of acquired resistance in bacteria [100]. QacA exhibits resistance against various cationic lipophilic antibacterial agents, particularly divalent ones [119].
In general, the MFS proteins are integral membrane proteins with two global structural bundles that are arranged in a distinctive MFS fold and asymmetrical functionally as well as structurally (Figure 4) [113, 120].
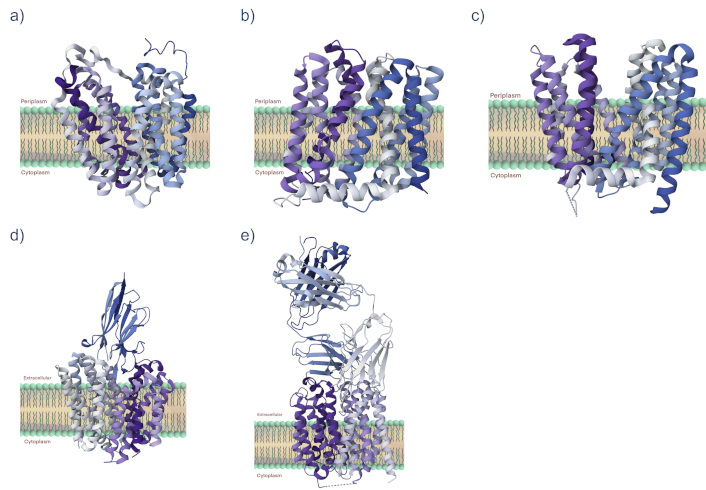
Protein structures of the major facilitator superfamily (MFS). Shown are three-dimensional ribbon structures for some proteins of the MFS for which a crystal structure has been determined by X-ray crystallography. For comparative purposes, we included (a) LacY (PDB 1PV6), the lactose permease, a symporter, from Escherichia coli [121], plus (b) MdfA (PDB 4ZOW) from E. coli [122, 123], (c) LmrP (PDB 6T1Z) from Lactobacillus lactis [124], (d) NorA conjugated to FAB36 (PDB 7LO8) from Staphylococcus aureus [125], and (e) NorC complexed with ICab3 (PDB 7D5P) from S. aureus [126, 127]. All protein structures are from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) https://www.rcsb.org/
During the solute transport mechanism, the two bundles are thought to interact mechanistically, involving conformational changes in structure and interaction as ion and antimicrobial substrates undergo translocation across the membrane through the MFS pumps [128]. The interhelical loop segments have been associated with gating, substrate specificity, and regulating conformational changes [129]. The transporters of the MFS share highly conserved amino acid sequence motifs [130]. These signature protein sequences have been demonstrated to be functionally important for specific aspects of antimicrobial efflux, whether for mediating conformational alterations, participating in molecular hinge structure operations, or speculatively coupling secondary energy to the active translocation of antimicrobial agents across the membrane [131]. The MFS transporters that confer antimicrobial efflux via ion-drug antiport are similar in protein structure, share similar sequences, harbor highly conserved signature sequence motifs A and C, and are evolutionarily related, predicting that these efflux pumps have a common ancestral origin [132–134]. These observations suggest that the MFS efflux pumps operate by a common mechanism of antimicrobial and ion translocation across the membrane.
As the MFS proteins mediate antimicrobial transport across the membrane, the drug binding sites in the channel are thought to be accessible on either side of the membrane depending on whether transport proceeds as symport or antiport, a process known colloquially as the “rocker-switch” or the so-called alternating-access mechanism [135]. Regarding antimicrobial efflux, the alternating access process involves the drug binding sites facing the cytoplasm in the resting pump structure and undergoing a reorientation during energized transport to mediate the binding site in an outward-facing configuration [106, 136].
Transport catalysis involving antimicrobial efflux in transport proteins of the MFS involves several putative stages. These transport systems are driven energetically by respiration-derived ion gradients, as proposed by Mitchell [137], supported by decades of investigations, summarized previously (Figure 5) [138, 139]. In the first step of the antimicrobial efflux catalysis mechanism, starting with a proton-bound MFS antimicrobial efflux pump, the binding site for the substrate (drug) faces outward [140]. Step two involves a respiration-driven proton binding to the MFS efflux pump, which enters an occluded stage devoid of its antimicrobial substrate [141, 142]. In step three, a molecular hinge mechanism formed by residues of the conserved motif C in helix five facilitates the formation of an inward-facing configuration, a process called switching, openly exposing the substrate (antimicrobial agent) binding site towards the cytoplasmic side of the membrane [114, 133, 137, 143]. Step four is characterized by antimicrobial binding to the MFS efflux pump and the release of ions from the occluded pump [144, 145]. In stage five, substrate binding drives the molecular hinge-mediated switching mechanism to form a configuration that is occluded by the bound antimicrobial agent [146–148]. During the sixth stage of the ion-driven drug efflux process (antiport), the antimicrobial agent is released to the outside of the cell. Protons bind to the empty, occluded efflux pump, leaving the MFS pump in an outward-facing configuration and ready to start the catalytic cycle once again [124, 141].
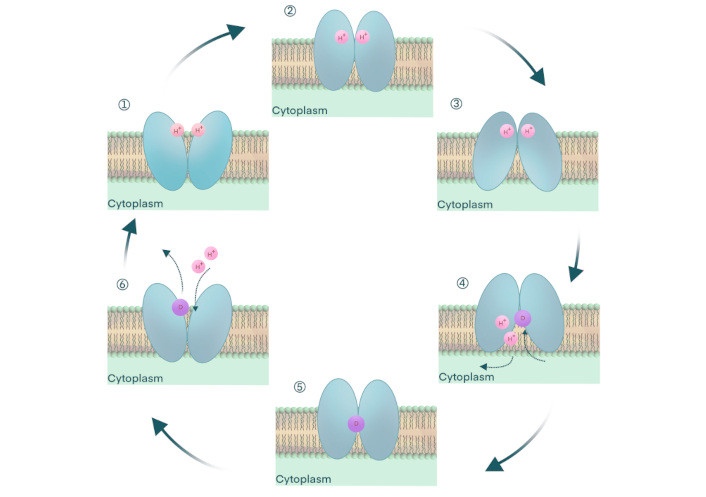
Antimicrobial efflux by ion-drug antiporters of the major facilitator superfamily (MFS) of proteins. The ion-driven antimicrobial agent efflux from cells with MFS transporters. (1) The efflux pump is open-outward, that is, empty of substrate and protons, and with its binding site for drug outward-facing. (2) Proton binds the empty MFS efflux pump and is occluded to prevent leakage. (3) A molecular hinge-mediated switching mechanism orients the drug binding site to an inward-facing open stage, a step mediated by the motif C, a highly conserved signature sequence. (4) The antimicrobial agent (substrate) binds the pump, and protons (ions) are released. The transporter is occluded at this stage, preventing undesired leakage. (5) Drug binding induces a switching mechanism by the molecular hinge to form a substrate-occluded conformation. (6) The pump is occluded after the substrate is released and protons (ions) have bound to the empty carrier. The MFS antimicrobial efflux pump is ready to start the translocation catalysis once again. This figure is adapted from Lekshmi et al. [99] (CC BY 4.0)
In general, the catalytic machinery involved in mediating antimicrobial transport across the membrane by the multidrug efflux pumps of the MFS represents suitable targets for modulating drug and MDR in bacterial pathogens [149], especially for the serious pathogenic strains of S. aureus [150, 151]. The development of efflux pump inhibitors (EPIs) targeting the MFS transporters has been a focus of investigation for decades [152, 153]. Such modulation of multidrug efflux in pathogenic bacteria has the potential to restore the clinical efficacy of chemotherapeutic agents that are compromised by resistance determinants in clinical pathogens [154]. Toward this, we discovered that an active principle in cumin, an aldehyde compound from the spice plant Cuminum cyminum, specifically targeted and inhibited antimicrobial transport in the MFS multidrug efflux pump LmrS from S. aureus [155]. In the same study, we also found that combinations of cumin extract and various antimicrobial agents acted synergistically to inhibit the growth of bacteria harboring over-expressed levels of LmrS, making this pump and other homologs suitable targets for modulation in different pathogens [155–157]. More recently, it was demonstrated that calcium ions enhanced the transport of antimicrobial agents by LmrS and that such efflux could be inhibited by calcium chelators [158]. Other investigations showed that the antimicrobial peptide plantaricin, PlnA, from Lactobacillus plantarum could bind specifically to MFS efflux pumps such as LmrS, MepA, and NorA and inhibit efflux of the antimicrobial agent ciprofloxacin, a known substrate of NorA [159]. Numerous natural compounds such as plant alkaloids, terpenes and terpenoids, lignins, flavonoids, essential oils, and coumarins and their semisynthetic derivatives modulate the NorA efflux pump and have been used as lead compounds to develop effective inhibitors of this MDR pump [160]. However, to date, no EPIs have been approved for clinical use.
Microorganisms that are causative agents of serious bacterial infection are increasingly frequent in the clinical setting. Morbidity and mortality rates are further exacerbated by antimicrobial resistance determinants harbored by clinical pathogens, especially those causative agents that are MDR [45]. In particular, MDR S. aureus strains are especially worrisome as such pathogenic strains are potentially untreatable, leading investigators to predict significantly enhanced mortality rates [161]. Global public health is facing a tremendous challenge in dealing with bacterial antimicrobial resistance. It was estimated to have caused almost 5 million deaths in 2021, and 1.3 million lives lost in 2019 were directly due to antibacterial agent-mediated clinical resistance [53, 161]. According to a recent report using a systematic meta-analysis of clinical infection data, it is predicted that 10 million deaths could occur annually by the year 2050 due to antimicrobial-resistant bacteria [161]. Currently, our ability to control MDR bacteria such as S. aureus is limited by the narrow treatment options involving very few antibiotics since no new class of anti-staphylococcal antimicrobial has been discovered in the recent past. However, MRSA and VRSA are on the World Health Organization’s list of “high priority” organisms for the development of new therapeutics [162]. The whole genome and transcriptomic data on S. aureus can help identify novel drug targets for the development of new drugs. This approach should also consider the resistance mechanisms, such as efflux pumps, which can reduce the efficacy of such drugs by lowering their intracellular concentrations. However, lengthy clinical trials involving newer drugs and the ability of S. aureus to develop resistance have been severe setbacks to the discovery of new therapeutics. In this context, recent approaches by some investigators for testing clinically approved synthetic drugs as EPIs have yielded encouraging results. For example, Food and Drug Administration (FDA)-approved drugs pyrvinium and raloxifene enhanced the bactericidal activity of ciprofloxacin on S. aureus and reduced the rate of resistance development through point mutations [163]. Pyrvinium is an anti-helminthic drug, while raloxifene is used to treat postmenopausal osteoporosis and reduce the risk of invasive breast cancer in post-menopausal women. Similarly, nilotinib, a tyrosine kinase inhibitor, has been shown to inhibit the NorA efflux pump, reduce the MIC of ciprofloxacin by two-fold, and reduce biofilm formation [164].
Pathogenic strains of S. aureus possess a variety of antimicrobial resistance mechanisms, many of which are determinants of MDR [45]. One of the most important of these resistance mechanisms involves the active export of multiple structurally distinct antimicrobial agents from the cells of S. aureus. In particular, secondary active efflux pumps of the MFS are prominently associated with bacterial MDR, sharing sequence, structure, and mechanistic characteristics of active drug transport. As such, these drug transporters and other resistance systems are active targets for modulation and potential restoration of clinical efficacy to alleviate and prevent rising morbidity and mortality.
Future work entails addressing these transport systems by studying the molecular physiology of antimicrobial efflux across the bacterial membrane to discover modulators and deliver them to individuals suffering from infection with S. aureus. While much of the catalytic steps involved during antimicrobial efflux have been characterized, it is nevertheless unclear how these transporters mediate the translocation of structurally dissimilar antimicrobial substrates and exclude undesirable leakage of ions or other molecules. Another focus of investigation that holds promise lies in examining the molecular mechanisms of energy transduction, which drive antimicrobial efflux. It is anticipated that knowledge of these energetic systems will lend themselves as modulation targets for circumventing bacterial resistance to antimicrobial agents.
Presently, it is poorly understood why the array of EPIs that have been discovered are not yet clinically available for the treatment of bacterial infections [94, 165]. It remains unclear why translational medicine has not been available for modulators of bacterial MDR, especially for pathogenic strains of S. aureus. Future efforts towards this particular area of clinical investigation are strongly encouraged.
12-TMS: 12-transmembrane segment
ATP: adenosine triphosphate
CA: community-associated
CC45: clonal complex 45
D-Ala-D-Lac: D-alanyl-D-lactate
EPIs: efflux pump inhibitors
HA: hospital-acquired
hetero-VRSA: heterologous-vancomycin-resistant Staphylococcus aureus
LA: livestock-associated
MDR: multidrug-resistant
MFS: major facilitator superfamily
MIC: minimal inhibitory concentration
MLST: multilocus sequence typing
MRSA: methicillin-resistant Staphylococcus aureus
PBP2a: penicillin-binding protein 2a
SCCmec: staphylococcal cassette chromosome mec
STs: sequence types
VISA: vancomycin-intermediate Staphylococcus aureus
VRSA: vancomycin-resistant Staphylococcus aureus
The authors thank Stephanie Luna, Aida Maamah, and Cecily Yandell for their helpful comments.
EA, MA, KFB, AOA, and CKD: Investigation, Writing—original draft, Writing—review & editing. ML: Investigation, Writing—original draft, Writing—review & editing, Conceptualization, Investigation, Validation, Supervision. SK: Conceptualization, Investigation, Writing—original draft, Writing—review & editing, Validation, Supervision. MFV: Conceptualization, Investigation, Writing—original draft, Writing—review & editing, Validation, Supervision, Funding acquisition. All authors read and approved the submitted version.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
The investigations from our laboratories and reported and considered in this review are supported in part by Faculty Research and Instructional Development (FRID) grants awarded by Eastern New Mexico University (ENMU) and research grants awarded by the National Institute of the General Medical Sciences [P20GM103451] from the National Institutes of Health, and the U.S. Department of Education, a Hispanic-Serving Institutions-Science, Technology, Engineering, or Mathematics and Articulation (HSI-STEM) Program [P031C110114]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
© The Author(s) 2025.
Open Exploration maintains a neutral stance on jurisdictional claims in published institutional affiliations and maps. All opinions expressed in this article are the personal views of the author(s) and do not represent the stance of the editorial team or the publisher.
Copyright: © The Author(s) 2025. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Irina Tiganova ... Yulia Romanova
