Affiliation:
1Department of Plant, Food and Environmental Sciences, Faculty of Agriculture, Dalhousie University, Truro, Nova Scotia B2N 5E3, Canada
ORCID: https://orcid.org/0000-0002-7742-347X
Affiliation:
1Department of Plant, Food and Environmental Sciences, Faculty of Agriculture, Dalhousie University, Truro, Nova Scotia B2N 5E3, Canada
2Natural and Applied Sciences, Hezekiah University, Umudi, Nigeria
Email: chijioke.emenike@dal.ca
ORCID: https://orcid.org/0000-0003-4306-0049
Explor Foods Foodomics. 2024;2:391–407 DOI: https://doi.org/10.37349/eff.2024.00043
Received: November 09, 2023 Accepted: May 16, 2024 Published: July 30, 2024
Academic Editor: Guoliang Li, Shaanxi University of Science and Technology, China
The article belongs to the special issue Food Authenticity and Emerging Challenges of Novel Food
The extraction, separation, and purification of dietary proteins from a variety of food sources are crucial for their targeted use in food applications. To achieve this, proteins should be effectively separated from non-protein components such as cell wall structures, polysaccharides, and lipids. Traditional protein purification methods can be time-consuming, highlighting the need for automated, cost-effective, and sustainable alternatives. This comprehensive review critically assesses various protein purification instruments from an analytical perspective, weighing their advantages and disadvantages. The methods under evaluation include ultrafiltration, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), fast protein liquid chromatography (FPLC), high-performance liquid chromatography (HPLC), ultra performance liquid chromatography (UPLC), and microfluidic chips. Among these, FPLC stands out as an affordable and efficient technique that allows for high protein recovery. However, HPLC and UPLC provide faster results but may denature proteins, leading to lower recovery rates. Ultrafiltration is a cost-effective and straightforward method that doesn’t require complex equipment. Microchip-based approaches are emerging as innovative techniques for rapidly analyzing small samples. While SDS-PAGE is user-friendly, it denatures proteins, particularly those linked to other biomolecules. The choice of the most appropriate instrument depends on factors such as cost, energy efficiency, processing time, the characteristics of the target protein, desired outcomes, protein recovery, and resource availability. By critically examining these analytical instruments for protein purification, this review aims to assist researchers and practitioners in selecting the most suitable method for their specific needs, ultimately promoting efficient and successful protein purification endeavors in the field of food science and technology.
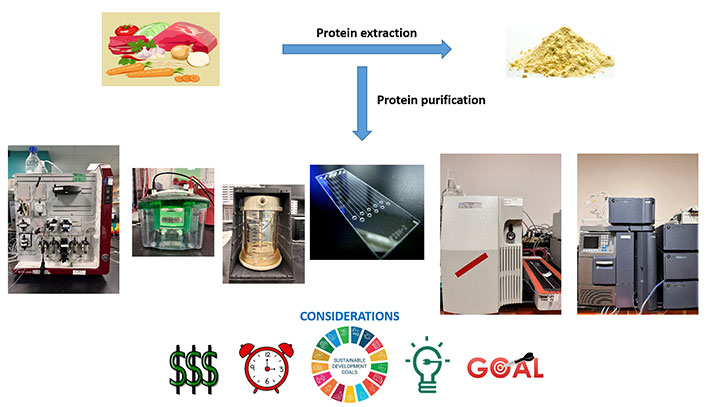
Protein separation methods, considerations, and goals
Note. Set of meat and vegetables vector image by tashh1601; protein powder by iStock.com/smirart; sustainable development goals by irinanaz, 123RF
Dietary proteins are recognized as essential components of the human diet, satisfying the body’s nitrogen requirements through the supply of both essential and non-essential amino acids [1]. Proteins, along with peptides, nucleotides, carbohydrates, and lipids, play a pivotal role in supporting growth, maintaining overall health, and providing energy and nutrients [2]. Moreover, proteins exhibit diverse functionalities, encompassing enzymatic activity, immunoprotection, and facilitation of biochemical transport across cellular membranes [3]. Beyond their primary roles, proteins, amino acids, and bioactive peptides offer numerous health benefits, including the reduction of risk factors associated with cardiovascular diseases, diabetes, cancer, obesity, and the improvement of bone health [4–7]. Animal-based proteins from sources such as meat, fish, eggs, and dairy products are widely consumed, while proteins derived from plants and algae, such as soybean protein, wheat protein, and seaweed proteins, are increasingly incorporated into meals as food ingredients [8].
The growing global population has exerted pressure on the agricultural production of protein resources, leading scientists to extensively research protein extraction from potential sources and explore methods to enhance them as food additives, nutritional supplements, and protein concentrates to meet the rising demand [2]. Natural proteins are often found in complex mixtures, with many exhibiting low yields and purity [9]. Obtaining a protein of interest necessitates extraction and subsequent purification, constituting a multi-step procedure. As detailed later in this paper, protein extraction is followed by the crucial steps of separation and purification.
The purification process aims to separate the target protein from non-protein components, isolating it among other constituents. To achieve optimal results from a desired protein, it is imperative to separate it from cellular components and other contaminants that may interfere with its structure and function [10]. In essence, for successful applications, proteins should possess sufficient purity and maintain a high signal-to-noise ratio. The purification step is widely regarded as the most laborious and time-consuming stage of the overall process [11]. Hence, the efficient utilization of analytical instruments for automation purposes holds significant importance [12]. Purification methods primarily rely on factors such as size, physicochemical properties, charge, binding affinity, and biochemical activity of the target protein(s). Additionally, the selection of a purification method depends on the specific research goals, feasibility, cost, sustainability, and other relevant factors.
This review delves into the practical aspects of widely used analytical instruments for protein purification from an analytical standpoint. We evaluated the instrumental advancements and effectiveness of SDS-PAGE, FPLC, HPLC, ultra performance liquid chromatography (UPLC), and microfluidic chips. This aims to provide researchers engaged in protein studies with comprehensive knowledge to select suitable purification methods for their work, considering the final objective, precision, feasibility, final quality, cost-effectiveness, time efficiency and sustainability.
Ultrafiltration (UF) plays a crucial role in protein separation and macromolecule concentration, primarily relying on differences in molecular size [13]. However, various factors, including solution pH, ionic strength, and system hydrodynamics, have been recognized for their impact on the separation process [14, 15].
Traditionally, UF relies on membrane performance, encompassing factors like selectivity and fouling properties. The pore size, structure, and material of the membrane significantly influence the separation process [16]. Furthermore, feed solution conditions, such as pH and ionic strength, process operating conditions, including flow rate, hydrodynamics, and flow orientation (normal versus tangential flow), also play a role in the overall performance [17].
To address these challenges and artifacts, stirred cell devices have been developed for UF. These devices come in various sizes, with capacities ranging from 3 mL to 400 mL. The device features a circular membrane disc, and fluid flow is facilitated by a mounted magnetic stir bar [18]. For protein concentration, gas pressure is directly applied to the UF cell. Solutes above the membrane’s molecular mass (MW) cut-off are retained within the cell, while water and solutes below the cut-off pass into the filtrate and out of the cell. Gentle magnetic stirring minimizes concentration polarization and shear denaturation [16].
UF offers the potential for cost-effective, high-throughput protein purification with high resolution for larger batches [19]. Notably, UF is suitable for processing biological samples, as it operates under mild conditions, preventing protein denaturation, deactivation, and degradation. Compared to chromatography, UF is cost-effective, scalable, and straightforward to implement. UF is widely used for protein concentration, solvent removal, buffer exchange, desalting, and virus elimination. It has also been explored for the fractionation of complex protein mixtures for food and biotechnological purposes, although commercialization remains a challenge due to limited selectivity in membrane systems [19].
Membrane fouling, attributed to broad pore sizes, bulk mass transfer, protein adsorption, protein deposition, and protein-protein interactions, is a primary hindrance to UF fractionation [20]. However, it is possible to fractionate similar MW proteins by adjusting solvent pH, salt concentration, and changing hydrodynamics, for instance, through gas sparging [21]. Membrane modifications, such as UV irradiation and protein pre-absorption, have shown promise in the separation of albumins and globulins [22]. The choice of membrane plays a critical role in the effectiveness of protein fractionation. Modified membranes can offer higher selectivity, permeate flux, and protein transmission compared to their original counterparts [23].
Furthermore, the compatibility of the membrane system with solution conditions such as pH and ionic strength is crucial for developing a commercial-scale purification unit. Successful fractionation of whey proteins and casein has been achieved when the membrane carries an opposite charge to that of peptides [24]. The highest protein transmission is typically observed at a solution pH close to the isoelectric point of the protein [17]. In the dairy industry, UF is employed to recover whey proteins released during cheese production [25]. Additionally, UF is a promising method for enriching algal proteins and isolating proteins and other macromolecules within the 1 kDa to 200 kDa range [25, 26]. For example, UF achieves 100% effective isolation of phycoerythrin protein from Grateloupia turuturu without denaturation [18].
The high throughput stirred cell (HTSC) is recognized for its efficiency in high-throughput screening and protein separation using UF [16]. HTSC allows six tests to be conducted simultaneously in parallel stirred cells, and it can be adapted for constant-pressure operation using a microfluidics control system. This system is amenable to statistical methods such as the design of experiments and requires substantially less sample volume compared to traditional Amicon® stirred cells. The collection of permeate samples in 96-well microplates facilitates integration with analytical instruments for further analysis [26].
Column and membrane chromatography are well-established methods for protein purification from complex feedstocks and mixtures, albeit with variations in application scale. While column chromatography may suffer from drawbacks such as fouling, poor bed compressibility, and slow flow rates, membrane devices exhibit comparable adsorption capacity and high scalability, in addition to being environmentally friendly and non-thermal [27]. Stirred cells offer ergonomic advantages, ease of assembly, quick connectors for tubing, integrated safety features, and autoclavability [14].
In summary, achieving high-resolution protein fractionation using UF requires fine-tuned conditions and process optimization, which can be time-consuming and resource intensive. Novel techniques, including HTSC and other advanced approaches like injection UF, carrier phase UF (CPUF), and high-performance tangential flow filtration (HPTFF), are being explored as competitive alternatives to chromatographic protein separation, potentially saving time, equipment, and resources [28].
SDS-PAGE is a well-established technique for separating proteins and peptides based on their size. The level of fractionation in SDS-PAGE is influenced by several factors, including MW, the length of the polypeptide chain, higher-order protein folding, post-translational modifications, and other factors affecting a protein’s ability to migrate in an electric field [29]. SDS, a chemical surfactant with detergent properties, plays a crucial role in this process. It disrupts secondary and tertiary protein structures, resulting in polypeptide chains that are negatively charged. When subjected to a voltage difference across a polyacrylamide gel, these charged proteins migrate according to their MW [30].
The Laemmli system, which utilizes a discontinuous buffer system, is widely employed for SDS-PAGE. This system involves pH 8.3 buffer for gel running, pH 6.3 buffer for the stacking gel, and pH 8.8 buffer for the resolving gel [31]. SDS-PAGE can effectively separate proteins with MW ranging from 5 kDa to 250 kDa. However, glycosylated proteins may not migrate according to their MW, leading to sizing precision variations of approximately 2–10% [32].
Proteins of interest can be excised based on their position relative to the standard proteins with well-defined MW and subjected to mass spectrometry (MS) for further analysis. Alternatively, the fractionated patterns can be transferred to a nitrocellulose membrane via western blotting for identification [29].
SDS-PAGE can be performed in two main formats: one-dimensional (1D) and two-dimensional (2D). While 1D SDS-PAGE separates proteins solely based on size, 2D gel electrophoresis (2D-GE) combines MW and the isoelectric point to achieve better resolution, making it a preferred choice in proteomics applications [33, 34]. Staining or scanning “empty” immobilized pH gradient strips after SDS-PAGE is advised as traces of protein can be detected, suggesting a potential increase in SDS concentration in the equilibration buffer might be necessary. Fluorescent dyes, offering sensitivity comparable to silver staining but better quantification, eliminate the need for fixing and staining, especially effective in cysteine labeling for minimal protein samples in 2D electrophoresis [35]. 2D-GE is the primary separation tool in proteomic analysis, crucial for comparative studies of protein expression under varying conditions, aiding in applications such as drug design and biomarker discovery. The integration of 2D-GE with other analytical techniques and the advancement of bioinformatics are expected to enhance proteomic research and set new standards in its applications [36]. α zein protein was efficiently isolated from maize using an eco-friendly method with 95% ethanol, and its homogeneity was analyzed using advanced techniques like matrix-assisted laser desorption/ionization (MALDI)-TOF-MS and 2D-GE, confirming the presence of distinct protein bands and spot groups. The MALDI mass spectra from these analyses provided marker ion signals characteristic of α zein protein, crucial for confirming its presence in hybrid corn products [37]. In cases where the target protein band is small and barely visible, western blot staining can be performed using readily available antibodies [38]. R-phycoerythrin, a fluorescent protein purified from red algae, is efficiently separated, and purified using chromatography, typically followed by SDS-PAGE for polypeptide analysis. The key distinction between native PAGE and SDS-PAGE is the use of SDS as an anionic detergent in the latter. In native PAGE, proteins are prepared in a non-denaturing and non-reducing buffer, allowing the secondary, tertiary, and quaternary structures, as well as charge, to influence protein migration [39].
Native PAGE at a neutral pH can separate proteins based on two modes. If the gel exhibits a sieving effect, then the separation is based on protein size (hydrodynamic diameter, MW), i.e., on the same principle as in the SDS-PAGE. If the gel serves only as an anticonvective medium gel pores are larger than the protein size (hydrodynamic diameter), then, the separation is based on different mobilities, i.e., charge/size (charge/hydrodynamic radius, charge/MW) ratio [40]. For the native PAGE, often performed in the form of pore-gradient PAGE. A short RON2 peptide (RON2) that binds to hydrophobic pocket on AMA1 parasite protein forming AMA1-RON2 complex is a key candidate for malaria vaccination, showing exceptional efficacy in preclinical studies. For clinical trial preparation, a native PAGE method along with western blotting, MS, and capillary isoelectric focusing (IEF) has been developed to accurately characterize and ensure the stability and integrity of the complex [41]. A significant advantage of native PAGE is that proteins can be recovered in their original state after separation [42]. This technique is relatively high-throughput and offers protein stability [43]. In contrast, SDS-PAGE denatures proteins into monomers, and protein mobility depends solely on MW due to the uniform charge-to-mass ratio introduced by SDS. While SDS-PAGE is effective for protein purification, separation, and MW determination, protein recovery is relatively low [42].
2D gel electrophoresis offers advantages such as high loading capacity, multiple wells for simultaneous sample analysis, scalability, and low complexity [33]. However, it has limitations, including base composition affecting fragment mobility, challenges visualizing bands with low-concentration samples, and glycosylated proteins not migrating according to their expected MW. Additionally, glycoproteins and lipoproteins may not be fully coated with SDS, leading to inaccurate MW estimations and protein identifications [39]. While, SDS-PAGE is commonly used to estimate MW and protein abundances, it may suffer from poor reproducibility between gels, making comparisons between different gels challenging [32]. Despite its utility, SDS-PAGE can be laborious and time-consuming, and it may not be easily automated. However, it remains a valuable method, especially when resources are limited, and expensive equipment and software are not readily available. SDS-PAGE is a powerful tool for protein analysis, offering valuable insights into MW and protein abundance. While it has its limitations and may not be suitable for all situations, it remains a practical and accessible technique for preliminary research in protein analysis.
Advancements in capillary and microchip electrophoresis (CE and MCE) have significantly improved the analysis and separation of proteins, with a particular focus on innovative coating technologies and integration with MS. Techniques such as successive multiple ionic-polymer layers (SMIL) for capillary coatings offer a promising alternative to traditional coatings, demonstrating superior separation efficiency and repeatability. Dynamic coatings, despite their potential interference with MS detection, have seen advancements that mitigate previous drawbacks, enhancing their utility in protein analysis [44, 45]. Additionally, developments in microchip electrophoresis have revolutionized traditional western blotting, enabling rapid, high-throughput analysis with significantly reduced sample sizes. Single-cell western blotting techniques have emerged, allowing for the detection of cell-to-cell variations with high specificity. These advancements not only provide powerful tools for protein characterization but also offer greater efficiency, sensitivity, and throughput in protein research. The integration of these techniques with MS and the development of novel polymer materials for MCE further expand the capabilities of protein analysis, making these methodologies indispensable in the field of proteomics and beyond [46, 47].
Free-flow electrophoresis (FFE) has been extensively utilized for the preparative separation and purification of a wide array of biomolecules and bioparticles, ranging from small biologically active peptides and drug enantiomers to proteins, nucleic acid fragments, and cellular components. The technique’s ability to operate in mild, carrierless media preserves the biological activity of the substances being separated, making FFE an invaluable tool in biotechnological and pharmaceutical research. While chromatographic methods still dominate large-scale purification, FFE’s integration into multidimensional separation systems offers significant advantages for analyzing complex mixtures in proteomics, demonstrating its potential for future advancements in microscale analytical and preparative separations [48]. Continuous flow electrophoresis (CFE) has advanced significantly, offering enhanced resolution for protein and peptide separation, efficient processing of large sample volumes, and simultaneous collection of multiple components. Innovations in CFE methodology and instrumentation, particularly in FFE and micro-FFE (µFFE), have led to improved separation capabilities, including the use of novel sample injection techniques and integration with other analytical methods for comprehensive biological sample analysis. CFE is particularly advantageous for preserving sample biological activity due to its operation in mild buffers without solid supports, making it ideal for the separation of cells, cellular organelles, vesicles, membrane fragments, and DNA. The method’s continuous processing ability, combined with real-time monitoring and optimization, positions CFE as a powerful tool in proteomics and bioparticle separation, facilitating advanced research in biological and medical sciences [49].
Protein purification aims to isolate a target protein from a mixture of contaminants, relying on the unique attributes of proteins such as size, MW, charge, and affinity [50]. Recent technological advances have led to the development of high-throughput chromatography methods, with 2D SDS-PAGE and liquid chromatography (LC) followed by MS emerging as promising techniques for protein isolation, fractionation, and identification [51]. HPLC is a critical analytical technique for the detection, separation, and quantification of drugs, requiring extensive method development and optimization including sample pre-treatment, mobile phase composition, column selection, and detector configuration. Its widespread application in drug analysis across development, manufacturing, and research necessitates rigorous method validation and optimization, ensuring analytical methods are robust, precise, and in compliance with Good Manufacturing Practice (GMP) standards [52].
FPLC is a HPLC variant known for its high resolution, achieved through the use of columns with small diameters. FPLC is characterized by its high loading capacity, biocompatible aqueous buffer system, fast flow rates, and common stationary phases such as ion exchange, reverse phase, affinity, gel filtration, and more [53]. Originally developed for proteins, FPLC has also been extended to oligonucleotides and plasmids [50].
FPLC is prized for its reproducible results and a high degree of automation, featuring techniques such as auto-sampling, gradient control, and peak collection [54]. In particular, anion exchange is a common FPLC method for protein separation, and the use of small particle sizes in column packing enhances chromatographic resolution by minimizing peak broadening. Theoretical plate height (H) and theoretical plate number (N) are directly influenced by particle size, with smaller particle sizes resulting in reduced H and improved resolution. HPLC, on the other hand, requires high pressure (up to 400 bar) and employs hyro-organic solvents, limiting its applicability for low sample loadings [55]. To address these limitations, FPLC was introduced in 1982 by Pharmacia Fine Chemicals in Uppsala, Sweden [56]. Unlike HPLC, FPLC operates at lower pressure (typically below 5 bar) and boasts comparatively high flow rates (ranging from 1–5 ml/min). FPLC can effectively analyze samples as small as 5 mL, containing milligrams of protein units, and is also suitable for industrial-scale production involving liters of samples [53].
The ÄKTA FPLC system which a fully automated platform designed for research-scale protein purification, offering cost-effectiveness and approximately 30 times lower costs than HPLC for a single test. The cost of FPLC columns is nearly ten times lower than that of HPLC columns with similar attributes [57]. FPLC offers a variety of chromatography models, including ion exchange, chromatofocusing, gel filtration, hydrophobic interactions, and reverse phase [58, 59]. Column particle diameters are in the same range as those used in HPLC, but FPLC columns can handle higher protein loadings in comparison to HPLC [60].
FPLC utilizes a dynamic range of aqueous buffer systems as mobile phases, ideally suited for biomolecules like proteins while avoiding denaturation [61]. The flow rate is typically controlled by a positive displacement pump, and buffer composition can be altered by drawing fluids from different external reservoirs in varying proportions [57].
Among the various modes of FPLC, anion exchange and gel filtration are commonly employed for protein analysis [50]. FPLC’s operation at lower pressure safeguards the integrity of fragile biomolecules. The stationary phase of FPLC primarily consists of porous resins, often cross-linked agarose, available in various bead sizes and surface ligands tailored to specific applications [27].
One notable advantage of FPLC is its capability to elute and collect each separated protein for further analysis. The volume of collected fractions can be precisely regulated as needed [62]. The UV detector in FPLC can be coupled with additional detectors for conductivity and pH, facilitating real-time monitoring of sample elution and buffer properties. Collection of separated sample fractions can be carried out either manually or automatically [50].
Moreover, FPLC fractionation can be conducted at low temperatures (4°C) to prevent denaturation of target proteins. This technique has been successfully applied to separate and analyze human milk whey proteins [63], as well as the extraction and separation of antimicrobial proteins like lysozyme and lactoferrin from donkey milk [64]. In another application, FPLC was used to detect adulteration of pasteurized bovine milk with soymilk, effectively reducing the limit of detection from 5% to 1%, compared to the time-consuming SDS-PAGE method [65]. FPLC instruments are constructed from biocompatible materials, offering an alternative to stainless steel [66]. FPLC is an essential tool for protein purification and fractionation, particularly when preserving the viability of biomaterials is a critical consideration.
In summary, FPLC stands as a versatile and cost-effective method for protein purification and analysis, offering a wide range of applications and several advantages over traditional HPLC. It’s a valuable tool for various research and industrial purposes, with its biocompatibility and low-pressure operation making it particularly suitable for proteins and biomolecules.
HPLC stands as the premier technique for the analysis and purification of biomolecules, including proteins, and has played a pivotal role in peptide and protein analysis for the past two decades, contributing significantly to breakthroughs in biological and medical research [67]. HPLC analysis boasts remarkable features such as high reproducibility, selectivity, excellent resolution, and high throughput capabilities [3]. It enables the separation of closely related proteins and peptides, as well as distinct ones. Proteins interact with the chromatographic column in an orientation-specific manner based on their molecular characteristics, leading to different retention times [68].
The principle behind HPLC’s protein separation involves the interactions between proteins and the stationary phase. These interactions are based on biological function or chemical structure, and affinity chromatography is often used for the purification and separation of closely related protein mixtures [69]. Reversed-phase HPLC (RP-HPLC) relies on hydrophobic interactions arising from the charges and surface properties of the protein species. This technique mainly employs a hydrophobic column for separation [70]. RP-HPLC is popular for separating complex protein or peptide mixtures, but the extreme experimental conditions can affect the 3D structure of proteins, leading to denaturation. Consequently, RP-HPLC is not suitable when preserving proteins in their biologically active form is paramount [71].
HPLC is highly versatile for protein isolation, regardless of their source, whether natural or synthetic. It has been a successful tool for purifying proteins and peptides, especially with the advent of solid-phase peptide synthesis and recombinant DNA technology, which increased the production of proteins and peptides requiring high levels of purification [67]. The complexity of purification depends on the nature of the protein source and the degree of preliminary cleanup achievable. RP-HPLC plays a pivotal role in the initial analysis and final large-scale purification of synthetic peptides [70].
HPLC operates under high pressure conditions (3.99 × 107–4.82 × 107 Pa) to drive solutes rapidly through the column. While diffusion is low, resolution remains high. The purification time varies based on the sample mixture’s nature [12]. After purification, proteins can be brought into a solution containing only volatile compounds, facilitating freeze-drying for further applications [3]. However, the use of organic solvents can disrupt the native protein structure. The high pressure used in HPLC can denature proteins, rendering them biologically inactive. Nevertheless, HPLC is a technique renowned for its analytical capabilities and versatility [9].
HPLC can be applied to analyze proteins at scales ranging from micron to nanoscale to large industrial processes. Computer models are available for simulating peptide separation parameters, reducing the number of experiments needed for method development and enabling the simulation of conditions that may not be feasible in the laboratory [72]. Additionally, HPLC can be coupled with MS, making it a vital tool in proteomics due to its excellent resolving power, convenience, versatility, stability, precision, and reproducibility. High-performance LC-electrospray ionization-MS (HPLC-ESI-MS) analysis of whole human saliva identified 120 proteins and revealed differential abundance of specific proteins between type 1 diabetes (T1D) patients and controls. [73]. HPLC’s ability to handle diverse analyte types with high separation power and sensitive detection is a significant advantage. However, perceived limitations include the absence of a suitable universal detector, less separation efficiency compared to capillary gas chromatography (GC), and potential labor intensiveness for regulatory or quality control purposes [74].
HPLC has found applications in the isolation and purification of food proteins from various sources. For instance, a study utilized HPLC-UV with a dialysate membrane to identify 11 proteins in coconut milk, offering a feasible and straightforward method for purification and enabling the structure prediction of newly identified proteins [75]. Multiple records exist of various HPLC techniques successfully isolating and identifying food proteins from sources like seaweeds, fish, wheat, rice, corn, milk, and more [76–78]. For example, RP-HPLC was employed to examine gluten concentrations in various Triticum species, concluding that all species were immunogenic [79].
In conclusion, HPLC remains the gold standard for the analysis and purification of proteins, playing a pivotal role in various fields of biology and medicine. Its versatility, combined with the advancements in peptide synthesis and recombinant DNA technology, has made it an indispensable tool for isolating and purifying proteins, whether from natural or synthetic sources. However, it's essential to consider denaturation when applying HPLC to purify proteins, as extreme conditions can affect their biological activity.
UPLC, or ultra pressure/performance LC, represents an evolution of HPLC, capable of operating at much higher pressures, typically ranging from 1.03 × 108 – 1.51 × 108 Pa as compared to HPLC’s 4.82 × 107 Pa. This elevated pressure imparts UPLC with superior sensitivity, enabling the analysis of complex samples with lower analyte quantities when compared to HPLC [80]. UPLC offers a better flow rate (2–5 ml/min) and higher resolution, making it faster than HPLC. Its exceptional separation efficacy is largely attributed to the increased pressure [81].
Compared to HPLC, UPLC provides higher sensitivity and allows faster analyses, even at the pico scale, as it employs columns with particle sizes less than 2 μm. These small particles in the stationary phase enhance peak shape and column resolution, with no compromise on flow rate, presenting another significant advantage of UPLC [80]. UPLC is highly effective for the purification, separation, and quantification of proteins and peptides, yielding significantly improved results over conventional HPLC [82]. UPLC has notably accelerated proteomics studies, thanks to groundbreaking work by James Jorgenson and colleagues, who introduced the first commercial UPLC system, Waters Acquity UPLC, in 2004 [74]. This system, equipped with a binary pump with a pressure limit of 1.03 × 108 Pa, an autosampler, a photodiode array (PDA) detector, and columns packed with 1.7 μm hybrid particles, quickly gained recognition and earned its place as a transformative innovation in modern HPLC. UPLC is also compatible with MS and chromatography data systems for detection purposes [74]. It boasts remarkable efficiency, achieving around 190,000 plates in 10 min using a capillary column packed with 1.5 μm nonporous particles at a pressure of 4.06 × 108 Pa. UPLC’s high acquisition rate of approximately 100 plates per second and a fast injection cycle of about 20 seconds further contribute to its success [74]. UPLC offers lower system dispersion (5–20 μL at 4𝜎 bandwidths) due to improvements in injectors, narrower inner diameters in connection tubes (< 0.005 inches), and smaller UV detector flow cells. Other features include low gradient dwell volumes (0.1–0.4 mL for binary pumps and 0.4–0.8 mL for quaternary pumps), shorter autosampler cycle times (three injections per minute), faster detector responses, and accelerated data acquisition (more than 40 points per second), making UPLC ideal for high-throughput screening [83]. UPLC also excels in injecting very small sample volumes with high precision, such as 1 μL and offers fast separation, high-resolution analysis, rapid method development, and the ability to customize resolution and performance. This flexibility allows the adjustment of column lengths and gradient times, increasing throughput by 3–5 times compared to conventional HPLC while maintaining similar resolution. However, it’s important to note that the small dwell volumes can negatively affect UV detector noise, and factors like flow range, column oven size, and sample loop size should be considered potential issues [84].
In drug analysis, UPLC has proven more accurate than HPLC. For instance, a study examined laxative tablets containing a natural product called senna using both HPLC and UPLC. The active pharmaceutical ingredient (API) of the drug consists of at least eight components. The original HPLC method failed due to the close elution of certain APIs, but UPLC, with a 300-mm long ultra high performance liquid chromatography (UHPLC) column packed with 1.7 μm particles, successfully resolved all eight APIs, providing a more accurate potency assay for the drug product [85]. UPLC has found applications in maintaining food safety. For instance, it is instrumental in detecting the deceptive addition of cow milk to camel milk powder using bovine β-lactoglobulin as a marker, an emerging food safety problem [86]. It is also utilized in the analysis of biological samples and the characterization of cell extracts via methodologies using single columns or in a 2D-LC platform. UPLC’s shorter analysis time and quicker column equilibration make it ideal for rapid method development [83].
Presently, analysts employ a variety of techniques, such as immunoassays, electrophoresis, and polymerase chain reaction, to detect the presence of pork and beef in mixtures [87]. The origin of minced meat is determined using UPLC. While SDS-PAGE is a manual and low-throughput method and PCR carries a high risk of contamination [88], SDS-PAGE also necessitates MS for protein identification, adding an extra step to the process. To address these challenges, a UPLC method was developed for identifying the source of minced meat using myoglobin protein as a marker [80]. This method is effective in simultaneously distinguishing myoglobin from water buffalo, bovine, pig, horse, chicken, and ostrich. Additionally, a rapid UPLC procedure was created to detect pork and beef in pre-mixed raw beef burgers, even in trace amounts [89]. UPLC has proven successful in ensuring food safety. For instance, China faces a growing concern regarding economically motivated milk powder adulteration. Scientists have developed a reliable method for identifying the fraudulent addition of cow milk to camel milk powder using UPLC, with bovine β-lactoglobulin as a marker [86]. Furthermore, an enzymatically hydrolyzed Porphyra dioica seaweed protein extract was fractionated through semi-preparative RP-HPLC, and the peptides were identified using UPLC-tandem MS (UPLC-MS/MS). This analysis revealed 33 peptides with potential applications as health-enhancing ingredients [90]. Apart from these, UPLC can be coupled with high-temperature LC, 2D-LC, and superficially porous particle (SPP) columns according to the requirement for better results [91].
New users may face challenges when adopting UPLC due to its high pressure, low flow rate (less than 1 ml/min), and small compressibility of liquids [92]. Pumping liquid at very high pressure through a small particle column can generate friction, resulting in a phenomenon known as viscous heating. This effect can be problematic, especially when dealing with column particles smaller than 2 μm, system pressures exceeding 8.27 × 107 Pa, and heat-sensitive analyte retention times. Minimizing errors due to this effect is essential, particularly for new users. Additionally, a thorough understanding of various factors like void volumes, peak volumes, dwell volumes, instrumental dispersion, peak capacity, and troubleshooting can be advantageous for users. Thus, comprehensive training is required before operating the instrument [92].
Another challenge is the price of UPLC instruments, which can be 10–50% higher than conventional HPLC instruments. Additionally, method translation can be a challenge in industries that use UPLC. While UPLC methods may not yet be standardized, methods for assays are typically converted to HPLC methods in one of three ways: they can be used as-is in both HPLC and UPLC, back-translated to HPLC, or translated from HPLC to UPLC. Nonetheless, UPLC is more efficient in both method development and analysis compared to HPLC, which can be advantageous, especially in the pharmaceutical and food sectors, where analysts demand portable methods applicable to both HPLC and UPLC [74].
Microfluidic chips are innovative tools that integrate the functions of sampling, separation, enhancement, and detection of biological analytes, including proteins, within a compact chip [93]. This automated method offers portability, high efficiency, accuracy, and cost-effectiveness [94]. The ability to use multiple small, low-cost chips in parallel allows for high-throughput protein separation [95]. These chips leverage the unique properties of liquids at micro or nanoscales and often apply the principles of LC or electrophoresis [96].
Microfluidic chips typically feature microchannels that can be modified using various types of particles. For instance, a microfluidic chip based on chromatofocusing with anion exchange particles successfully separated a protein mixture composed of two proteins with close isoelectric points (pI) based on their pI values [97]. These microchips have seen significant improvements in structure and function over time, with innovations such as cross-intersection channels designed to drain ion-depleted buffer from analytical channels and reintroduce running buffer, avoiding issues with electro-pre-concentration [98]. Another study introduced an arrayed pH gradient with 80 parallel separation channels using a two-layer open IEF microfluidic chip to achieve high-throughput results [99].
The surface of microfluidic chips can be modified with substances like polyethylene glycol diacrylate to minimize non-specific protein adsorption while enhancing the detection of protein biomarkers related to human and animal health [100]. Microfluidic chips have found applications in protein analysis with various detection methods. For instance, Agilent 2100 bioanalyzer with epi-fluorescent detection was used to determine proteins in soybean flour, proving more efficient than SDS-gel capillary electrophoresis [101]. It has also been employed to study fish protein expression changes induced by aquaculture by denaturing proteins on-chip using an Agilent protein reagent kit [102]. Furthermore, amino acids in various beverages have been analyzed using this technique, offering fast results, with the entire analysis taking less than 10 min, including sample preparation and fluorescence labeling [103]. It is also effective for food allergen detection [104].
Throughout the last century, microfluidic platforms have witnessed immense improvements, with a focus on precise chemical analysis of biomolecules, including proteins. One of the key advantages of this technology is the ability to use extremely small sample volumes as analytes. Moreover, high reproducibility, automation, integration, and precision are among the benefits derived from microfluidic chips [105]. Their high protein selectivity, sensitivity, and compatibility with miniature devices make this technique well-suited for complex biochemical screening, proteomics, and bioprocess development [97]. Notably, microfluidic chips are user-friendly, effective, and more environmentally friendly, making them a valuable tool for protein analysis in food samples.
In summary, protein separation from diverse plant and animal food sources traditionally demanded substantial resources, labor-intensive efforts, and significant energy consumption. The imperative shift toward automation in protein separation and fractionation necessitates analytical instruments that excel in various critical aspects such as cost-effectiveness, time efficiency, energy conservation, extraction effectiveness, protein recovery, and scalability.
The UF presents itself as a compelling method due to its economic nature and mild operating conditions, eliminating the need for toxic chemicals. It stands out for its ability to prevent protein denaturation, deactivation, and degradation, particularly when compared to conventional chromatography methods. Meanwhile, SDS-PAGE exploits protein size to separate proteins, although it struggles with relatively low protein recovery. However, its significant loading capacity and simplicity render it an effective choice for certain applications, although it may not be ideal for glyco and lipoprotein separations.
Chromatography, a versatile protein separation technique, offers various modes like FPLC, HPLC, and UPLC. FPLC’s low-pressure operation significantly reduces costs, with a per-test expense around 30 times lower than HPLC, accompanied by a substantially more cost-effective column. Thus, FPLC emerges as a fully automated, cost-effective method with scalability potential to industrial levels. HPLC, known for its high-pressure operation (3.99 × 107–4.8 × 107 Pa) and superior resolution, presents an alternative choice. UPLC, with its capacity to handle even higher pressures (1.03 × 108–1.51 × 108) and enhanced sensitivity, offers the advantage of a faster and more efficient process, albeit at the cost of lower protein recovery.
An innovative approach to protein separation is the microfluidic chip, which provides the capability for parallel protein separation and detection within a compact, portable device. With its automated operation, high efficiency, precision, and cost-effectiveness, it represents an ideal method for protein identification and detection, particularly for point-of-care and resource-constrained settings.
Selecting the optimal method for protein separation hinges on specific requirements, desired outcomes, the nature of the target protein, and the sustainability of the chosen technique. Each method exhibits unique strengths and limitations, making it crucial to align the selection with the specific demands of the task at hand.
2D: two-dimensional
2D-GE: two-dimensional gel electrophoresis
API: active pharmaceutical ingredient
CFE: continuous flow electrophoresis
FFE: free-flow electrophoresis
FPLC: fast protein liquid chromatography
GC: gas chromatography
HPLC: high-performance liquid chromatography
HTSC: high throughput stirred cells
IEF: isoelectric focusing
LC: liquid chromatography
MS: mass spectrometry
MW: molecular mass
RP-HPLC: reversed-phase high-performance liquid chromatography
SDS-PAGE: sodium dodecyl sulfate-polyacrylamide gel electrophoresis
SPP: superficially porous particle
UF: ultrafiltration
UPLC: ultra performance liquid chromatography
AMW: Conceptualization, Data curation, Writing—original draft, Writing—review & editing. CE: Conceptualization, Supervision, Writing—review & editing.
The authors declare that there are no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
All the data are incorporated in the review article.
Not applicable.
© The Author(s) 2024.
Copyright: © The Author(s) 2024. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Bernhard Thalhamer ... Wolfgang Buchberger
Yanmei Zhu ... Haiyan Fu
Adindu O. Onyeodili ... Raquel P.F. Guiné
