Affiliation:
1Department of Pulmonology and Respiratory Medicine, Faculty of Medicine, Hasanuddin University, Makassar 90245, Indonesia
2Department of Pulmonology, Dr. Wahidin Sudirohusodo Hospital, Makassar 90245, Indonesia
ORCID: https://orcid.org/0009-0007-3155-1680
Affiliation:
1Department of Pulmonology and Respiratory Medicine, Faculty of Medicine, Hasanuddin University, Makassar 90245, Indonesia
2Department of Pulmonology, Dr. Wahidin Sudirohusodo Hospital, Makassar 90245, Indonesia
ORCID: https://orcid.org/0000-0003-0108-1554
Affiliation:
1Department of Pulmonology and Respiratory Medicine, Faculty of Medicine, Hasanuddin University, Makassar 90245, Indonesia
2Department of Pulmonology, Dr. Wahidin Sudirohusodo Hospital, Makassar 90245, Indonesia
ORCID: https://orcid.org/0000-0001-9012-7475
Affiliation:
1Department of Pulmonology and Respiratory Medicine, Faculty of Medicine, Hasanuddin University, Makassar 90245, Indonesia
2Department of Pulmonology, Dr. Wahidin Sudirohusodo Hospital, Makassar 90245, Indonesia
Email: irawatydjaharuddin@unhas.ac.id
ORCID: https://orcid.org/0000-0002-5240-4950
Affiliation:
1Department of Pulmonology and Respiratory Medicine, Faculty of Medicine, Hasanuddin University, Makassar 90245, Indonesia
ORCID: https://orcid.org/0009-0005-2249-4114
Affiliation:
1Department of Pulmonology and Respiratory Medicine, Faculty of Medicine, Hasanuddin University, Makassar 90245, Indonesia
ORCID: https://orcid.org/0000-0003-2285-7286
Affiliation:
1Department of Pulmonology and Respiratory Medicine, Faculty of Medicine, Hasanuddin University, Makassar 90245, Indonesia
2Department of Pulmonology, Dr. Wahidin Sudirohusodo Hospital, Makassar 90245, Indonesia
ORCID: https://orcid.org/0000-0002-2148-4353
Affiliation:
3Department of Epidemiology, Faculty of Public Health, Hasanuddin University, Makassar 90245, Indonesia
ORCID: https://orcid.org/0000-0002-9692-6136
Affiliation:
4Faculty of Medicine, Hasanuddin University, Makassar 90245, Indonesia
ORCID: https://orcid.org/0000-0002-3944-9556
Affiliation:
4Faculty of Medicine, Hasanuddin University, Makassar 90245, Indonesia
ORCID: https://orcid.org/0000-0003-1179-5366
Explor Immunol. 2024;4:679–690 DOI: https://doi.org/10.37349/ei.2024.00166
Received: June 13, 2024 Accepted: September 23, 2024 Published: October 29, 2024
Academic Editor: Wenping Gong, The Eighth Medical Center of PLA General Hospital, China
The article belongs to the special issue Novel Vaccines development for Emerging, Acute, and Re-emerging Infectious Diseases
Aim: Assessing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) immunoglobulin G (IgG) antibodies among healthcare workers (HCWs) is crucial in understanding the durability of humoral immune response due to vaccination and the post-pandemic era. Therefore, this study aimed to evaluate IgG antibody seroprevalence, IgG levels, and long-term durability responses after the coronavirus disease 2019 (COVID-19) pandemic in HCWs.
Methods: The cross-sectional study recruited HCWs aged ≥ 18 years, vaccinated at least once, and without autoimmune diseases at Dr. Wahidin Sudirohusodo Hospital Makassar. Antibody titers against spike protein, nucleocapsid protein, and spike-nucleocapsid conjugate were quantified using dried blood spot (DBS) samples and multi-antigen profiling (xMAP) technology to measure seroprevalence.
Results: The results showed that there was a total of 103 participants with the majority being female (59.22%), ≤ 40-year-old (87.38%), nurses (56.31%), had no comorbidities (88.35%), and were infected with COVID-19 (70.87%). A significant proportion received three times vaccination (76.7%), while 102 of 103 (99.03%) showed seropositivity. IgG showed the highest mean antibody titers for spike protein [10,819.38 ± 2,554.23 mean fluorescence intensity (MFI)] in the first 6–12 months after the last vaccinations. Meanwhile, the IgG titer (MFI) showed a long duration over 30 months. Vaccine significantly influenced spike protein antibody levels, with the highest observed in the group receiving a combination of two inactivated vaccines followed by two messenger ribonucleic acid (mRNA) vaccines (p < 0.05).
Conclusions: HCWs showed a sustained humoral immune response to SARS-CoV-2 for over 24 months post-vaccination. The type and combination of vaccines administered were significantly correlated with the IgG antibody levels, suggesting the long-term efficacy of the vaccination regimens. The application of DBS and xMAP technology in the study facilitated the assessment of SARS-CoV-2 IgG responses, offering valuable insights into the dynamics of immune persistence following vaccination.
Coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was reported to induce unprecedented global health challenges following the initial discovery in Wuhan, China, in December 2019 [1]. After rapidly spreading across borders, the World Health Organization (WHO) declared COVID-19 a pandemic on March 11, 2020. As of June 9, 2023, the pandemic has resulted in 767,750,853 confirmed cases and 6,941,095 deaths worldwide, with Indonesia reporting 6,809,377 confirmed cases and 161,811 deaths [2–4]. In response to this crisis, vaccination has been identified as a crucial measure to control the spread of the virus. Indonesia has achieved a vaccination rate of 86.87% for the first dose and 74.53% for the second dose as of the same date, prioritizing vulnerable populations specifically healthcare workers (HCWs) [5, 6].
HCWs are at a heightened risk of contracting COVID-19 due to direct exposure to patients. Therefore, vaccination is not only beneficial for health but also instrumental in maintaining healthcare system functionality by reducing staff shortages and the risk of virus transmission to patients [7–9]. Immunoglobulin G (IgG) plays a crucial role in the humoral immune response to SARS-CoV-2 infection. Through several mechanisms, including pathogen neutralization, opsonization, and complement system activation, IgG antibodies are crucial for identifying and binding to the S1 domain of the spike protein, as measured by enzyme-linked immunosorbent assay (ELISA) in serum or dried blood spot (DBS) samples [10–12].
Given the limited data, particularly in Indonesia, regarding IgG levels among vaccinated HCWs, assessing SARS-CoV-2 IgG antibodies among HCWs is essential in understanding the durability of humoral immune response post-vaccination and post-pandemic. Therefore, this study aimed to evaluate IgG antibody seroprevalence and the levels after the COVID-19 pandemic in HCWs.
This was an observational, cross-sectional study, conducted at Dr. Wahidin Sudirohusodo Hospital Makassar in the eastern part of Indonesia. The participants included HCWs recruited from September–October 2023. The inclusion criteria were age ≥ 18 years, assigned as HCWs during the pandemic (doctor, nurse, pharmacist, laboratory analyst, radiologist, etc.), and vaccinated ≥ 1 time. Participants with a history of comorbidities such as coronary heart disease, congestive heart failure, renal insufficiency, chronic liver disease, uncontrolled diabetes mellitus, uncontrolled hypertension, immunocompromised, CNS diseases, or signs of acute SARS-CoV-2 infection were excluded.
In this study, comprehensive vaccination data were obtained from the digital vaccination card of each participant accessible through the mobile application ‘Satu Sehat’. This platform records the type of vaccines administered, such as inactivated (e.g., Coronavac, Biofarma), viral vector (ChAdOx1 nCoV-19), and mRNA vaccines (Moderna and Pfizer). HCWs at Dr. Wahidin Sudirohusodo Hospital received vaccinations based on availability or personal choice at various vaccination centers, allowing for a range of vaccine-type combinations among the participants.
This study used the Slovin sampling formula and within the sample population that matched the eligibility criteria, a total of 3,606 individuals were identified. A tolerance level for the margin of sampling error deemed acceptable was set at 10% (e = 0.10), resulting in a minimum sample size of 97.
DBS samples were collected from participants who met the sample criteria and subsequently stored on Whatman 3.0 protein saver cards. These samples were then air-dried at room temperature for 3–4 hours. After labeling, the DBS samples could be preserved in a refrigerator at 4–8 degrees (Celsius) [13]. The processing was carried out at the Virology Laboratory in Environmental Health Engineering and Disease Control Agency Makassar.
Subsequently, DBS samples were diluted, and centrifuged, using Tetracore® FlexImmArray™ SARS-CoV-2 Human IgG Antibody Test enabling the assessment of multi-antigens against spike protein, spike-nucleocapsid conjugate, and nucleocapsid protein. These components were quantified multi-antigen profiling (xMAP) technology by Luminex MAGPIX® XPON4-CEIVD (Austin, Texas) to determine seroprevalence [14]. Adapted from a recent source by Mazhari et al. [14] and Miesse et al. [13], the examination method is presented in Figure 1.
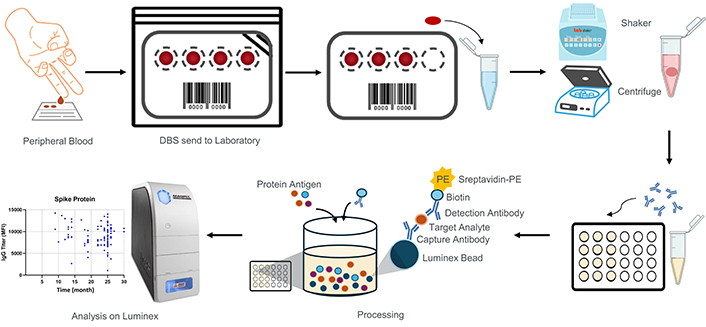
Sample collection and IgG analysis workflow. Dried blood samples were collected from the peripheral blood of participants and placed onto protein saver paper. In the laboratory, paper discs were punched out and placed into microtubes. These discs were eluted using buffer. Subsequently, the samples were processed with a fluorescence reporter (anti-human IgG-PE) and antigen protein. The Luminex xMAP technology was used for both qualitative and quantitative analysis of IgG levels. DBS: dried blood spot; IgG: immunoglobulin G; MFI: mean fluorescence intensity; PE: phycoerythrin
Initially, a 6 mm DBS punched from a protein saver paper with concentrated blood is placed into a 1.5 mL tube (Axygen, MCT-150-C), and 500 µL of sample dilution buffer is added. The hole puncher is cleaned after each punching to reduce the potential for cross-contamination. All samples are shaken at 1,200 rotation per minute (rpm) for 30 minutes at room temperature. The samples are then centrifuged for 8 minutes at a speed of 3,000 rpm or 8,000 rpm.
After finished DBS extraction, 10 µL of the 7-plex microsphere mixture was added to each well of the Tetracore plate. Subsequently, 50 µL of the DBS extraction solution from the 1.5 mL tubes was added to the corresponding wells according to a pre-made map. Following this, 50 µL of the ready-to-use Positive Control, Calibrator, and Negative Control was added to the designated wells as per the plate map. The plate was then sealed with an aluminum plate sealer and incubated in the dark at room temperature for 20 minutes while shaking at 800 rpm. The plate was washed 4 times with 200 µL of 1× assay wash buffer using the Luminex magnetic plate separator. After pipetting up and down, 25 µL of fluorescence reporter (anti-human IgG-PE) from the Tetracore Kit was added to each well. The plate was covered with an aluminum plate sealer and incubated at room temperature for 20 minutes while shaking at 800 rpm. The test plate was washed again 4 times with 200 µL of 1× assay wash buffer. Then, 100 µL of 1× assay wash buffer was added to each well of the test plate, covered with an aluminum plate sealer, and re-suspended for 5 minutes while shaking on the Luminex MAGPIX instrument at 800 rpm. Finally, the plate was analyzed using the Luminex MAGPIX®.
Categorical variables were summarised with counts and percentages before being compared using the Chi-square test. Furthermore, comparisons between means of continuous variables were performed using independent t-tests for two-group comparisons and one-way ANOVA for multiple-group comparisons. The ANOVA was followed by a post-hoc analysis using the Tukey test to determine specific group differences. Statistical significance was established at a p-value of < 0.05. IgG titer in MFI is presented schematically using GraphPad Prism 10.
Between September and October 2023, a total of 117 HCWs were invited from various departments at Dr. Wahidin Sudirohusodo Hospital in Makassar, Indonesia, to participate in the study. The departments included the Intensive Care Unit (ICU), Emergency Room (ER), Ward, Polyclinic, and Operation Room (OR). Among the 117 HCWs initially registered for screening, five were excluded due to not receiving any COVID-19 vaccination. Following this, 112 HCWs were interviewed to further verify eligibility. During this phase, two additional participants were excluded because the provided vaccination evidence was unclear and did not match specified requirements.
Blood samples were then collected from the remaining 110 HCWs. However, seven were deemed invalid due to issues such as high background noise and low bead count in the assays, leading to the exclusion from the study. Finally, 103 eligible HCWs were enrolled including 20 from the ICU, 25 from the ER, 19 from the Ward, 19 from the Polyclinic, and 20 from the OR. The detailed sampling process is shown in Figure 2, and the characteristics of the participants are summarized in Table 1.
Characteristics of the study participants
| Profile | Number (n = 103) | (%) | |
|---|---|---|---|
| Age (years) | ≤ 40 | 90 | 87.38 |
| > 40 | 13 | 12.62 | |
| Gender | Male | 42 | 40.78 |
| Female | 61 | 59.22 | |
| Body mass index | Underweight | 2 | 1.94 |
| Normal | 26 | 25.24 | |
| Overweight | 28 | 27.18 | |
| Obesity class 1 | 33 | 32.04 | |
| Obesity class 2 | 14 | 13.59 | |
| Occupation | Doctor | 34 | 33.01 |
| Nurse | 58 | 56.31 | |
| Other | 11 | 10.68 | |
| Work unit | Intensive care unit | 20 | 19.42 |
| Emergency room | 25 | 24.27 | |
| Ward | 19 | 18.45 | |
| Policlinic | 19 | 18.45 | |
| Operation room | 20 | 19.42 | |
| COVID-19 history | Infected | 73 | 70.87 |
| Non-infected | 30 | 29.13 | |
| Vaccination frequency (times) | 2 | 5 | 4.85 |
| 3 | 79 | 76.70 | |
| 4 | 19 | 18.45 | |
| Duration (months) | 6–12 | 6 | 5.83 |
| 13–18 | 16 | 15.53 | |
| 18–24 | 33 | 32.04 | |
| 25–31 | 48 | 46.60 | |
| Vaccine combination | 2 Inactive - 2 mRNA | 14 | 13.59 |
| 2 Inactive - mRNA | 78 | 75.73 | |
| Others | 11 | 10.68 | |
| Vaccine status | Full-Infected | 10 | 9.71 |
| Full-Non Infected | 9 | 8.74 | |
| Not Full-Infected | 63 | 61.17 | |
| Not Full-Non Infected | 21 | 20.39 | |
| IgG antibody | Seropositive | 102 | 99.03 |
| Seronegative | 1 | 0.97 | |
IgG: Immunoglobulin G; mRNA: messenger RNA; Full-Infected: vaccinated 4 times and confirmed COVID-19; Full-Non Infected: vaccinated 4 times and not confirmed COVID-19; Not Full-Infected: < 4 vaccinations and confirmed COVID-19; Not Full-Non Infected: < 4 vaccinations and not confirmed COVID-19
Among all 103 participants, 99% were seropositive and only one was seronegative as shown in Table 1. The seronegative one was shown by a male, ≤ 40 years old, male, Bugis tribe, Bachelor’s degree, obesity 1, worked in the operating room attendant as an anesthetist (in other categories), has no comorbidities, infected with COVID-19, received three vaccines and the last vaccine has been > 24 months.
The titer of spike protein (10,819.38 MFI) and spike-nucleocapsid conjugate (10,627.00 MFI) was highest in ‘shortest duration’ between the last vaccination and blood sample, which was 6–12 months. On the contrary, the nucleocapsid protein titer was highest in the ‘longest duration’ between the last vaccination and blood sample, which was > 24 months (3,195.73 MFI). The data are presented in Figure 3 (Table S1).
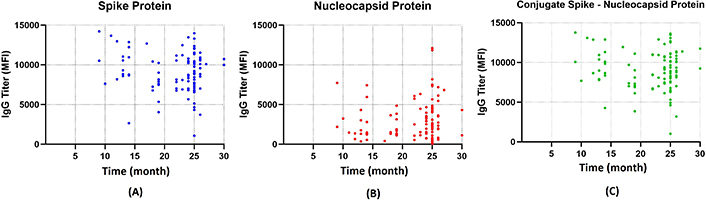
Durability of the antibody response. Scatter diagram illustrating the titers of IgG against SARS-CoV-2 in MFI units plotted against the duration from the last vaccination to the blood sample test (months). (A) Spike protein depicted as blue dots detected and sustained up to month 30. (B) Nucleocapsid protein detected tends to be lower compared to spike protein but still detectable up to month 30. (C) Spike-nucleocapsid protein conjugate. IgG: immunoglobulin G; MFI: mean fluorescence intensity
In this study, participants with the highest number of vaccination doses (4 times) gained the highest result of spike protein (10,219.21 MFI) and spike-nucleocapsid conjugate (1,000.26 MFI) titer among others. The opposite result was found in nucleocapsid protein titer, where the least was found in the 4 times group (2,281.84 MFI) although the result was not significant as shown in Figure 4.
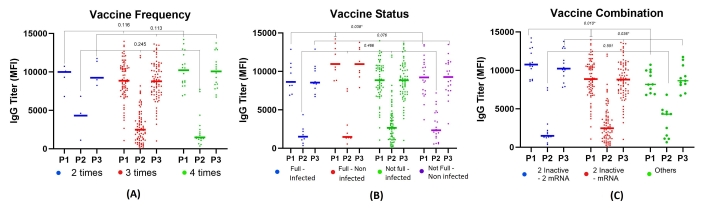
The titers of IgG based on vaccination frequency, status, and combination. (A) IgG titers based on vaccination frequency showed higher levels in the group vaccinated 4 times. (B) IgG titers based on the combination of vaccine doses and COVID-19 history, showing significantly higher mean titers in the group vaccinated 4 times and non-infected with COVID-19, with significant differences in spike protein (p < 0.05). (C) Mean IgG titers found highest in the group receiving a combination of 2 inactivated vaccines and 2 mRNA-based vaccines, with significance observed in spike protein and spike-nucleocapsid protein conjugate (p < 0.05). Full-Infected: vaccinated 4 times and confirmed COVID-19; Full-Non Infected: vaccinated 4 times and not confirmed COVID-19; Not Full-Infected: < 4 vaccinations and confirmed COVID-19; Not Full-Non Infected: < 4 vaccinations and not confirmed COVID-19. IgG: immunoglobulin G; MFI: mean fluorescence intensity; mRNA: messenger RNA; P1: spike protein; P2: nucleoprotein; P3: spike-nucleocapsid protein conjugate
Participants with complete doses of vaccination and non-infected COVID-19 had the highest result of spike protein (11,380.11 MFI) and spike-nucleocapsid conjugate (11,012.00 MFI) titer compared to other groups. The result of spike protein titer was significantly different. Meanwhile, the nucleocapsid protein was found to be higher in participants who had incomplete doses of vaccination but were infected with COVID-19 (p = 0.466) (Table S2, Figure 4, Figure S1).
Types of vaccination had a significant result of spike protein (p = 0.010) and spike-nucleocapsid conjugate (p = 0.036) titer. The highest result was in two inactive - 2 mRNA groups compared to the others as shown in Figure 4. Post hoc analysis was conducted to identify the differences in IgG among four different groups of vaccination status, as shown in Table S3. There were significant differences in the mean IgG spike protein and conjunction between ‘Fully Vaccinated, Non-Infected’ and ‘Not Fully Vaccinated, Infected’ (p < 0.05). On the other hand, there were significant differences in IgG spike protein among three groups in vaccine combination (p < 0.05), as well as spike-nucleocapsid conjugate ‘2 inactive - 2 mRNA’ group and ‘2 inactive - mRNA’ group (p < 0.05). As shown in Table S4, the statistical power of the analysis was determined to be 0.68 (alpha 0.05).
This cross-sectional study characterized the immune response of 103 HCWs and explored the relevant factors, such as duration and vaccination schema (frequency, status, and type of vaccine). Anti-spike (10,819.38 MFI) and spike-nucleocapsid conjugate (10,627.00 MFI) were found to be highest in the ‘shortest duration’ between the last vaccination and blood sample, which was 6–12 months. However, in the ‘most extended duration’ group (25–31 months), both were still detectable with 9,303.85 MFI and 9,303.85 MFI titers for anti-spike and spike-nucleocapsid conjugate, respectively. Based on a previous study, the anti-spike against SARS-CoV-2 could last up to 270 days [15]. Another study reported that the maximum duration of spike protein IgG antibodies was 744 days [16].
The study of IgG SARS-CoV-2 levels among vaccinated HCWs showed a significantly high seropositivity rate of 99.03%. This result is consistent with recent studies [17–19]. A systematic review and meta-analysis before vaccination reported an average IgG seropositivity of 8.6% across three continents including America, Europe, and East Asia. This comparison underscored the significant impact of vaccination on immune response [20].
Another aim of this study was to address the production of IgG antibodies in vaccinated HCWs who already received at least one step of vaccination. It was found that one out of 103 samples showed seronegative results. This might be because a short-term antibody response was induced, as reported for mild or asymptomatic infections by SARS-CoV-2 [21–23]. The mechanism of humoral antibody response due to the natural infection and vaccination is shown in Figure 5 [24, 25]. A study by Sugiyama et al. [26] in Japan also identified a seronegative participant under different conditions. Immunosuppressant therapy was not extensively screened in the study, and this factor is crucial for further investigation to understand the impact on vaccine-induced immunity [26]. Exploring the factors for the seronegative sample might be necessary, specifically in HCWs, for further study.
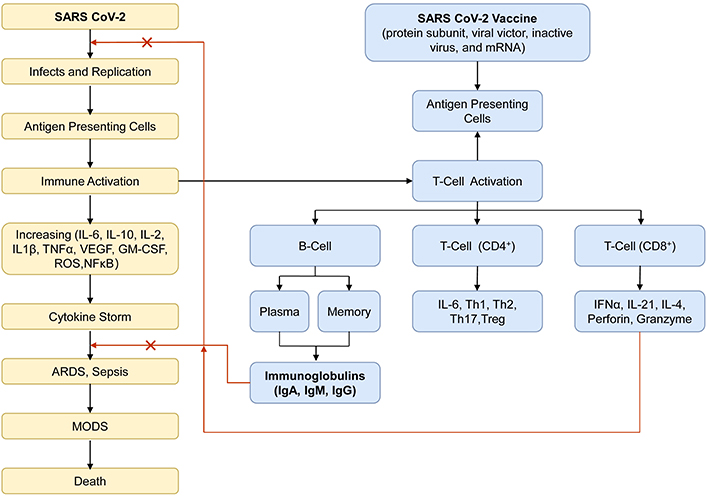
The mechanism of humoral antibody response. The schematics picture illustrates how natural infection with the SARS-CoV-2 virus and vaccination can affect the production of immunoglobulins through the activation of adaptive immunity, which functions as specific antibodies in preventing the severity of COVID-19. ARDS: acute respiratory distress syndrome; CD: cluster of differentiation; GM-CSF: granulocyte-macrophage colony-stimulating factor; IFN: interferon; Ig: immunoglobulin; IL: interleukin; MODS; multiple organ dysfunction syndromes; mRNA: messenger RNA; NF-κB: nuclear factor-κB; ROS: reactive oxygen species; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2; Th: T helper; TNF: tumor necrosis factor; Treg: T regulatory; VEGF: vascular endothelial growth factor
The high risk of pathogen exposure among HCWs due to regular interaction with patients might explain the elevated seropositivity levels, reflecting either repeated exposure or vaccination interventions [17]. Hossain et al. [20], found higher IgG SARS-CoV-2 antibody seropositivity rates among HCWs in direct patient contact compared to lower-risk groups. During a natural infection, the adaptive immune system responds, and by the time an antibody response is mounted, numerous cells have already been infected [24].
The use of vaccine combinations has been suggested to elicit a stronger, more durable antibody response than single-vaccine regimens. For example, administering a Moderna booster to individuals previously vaccinated with two doses of CoronaVac increased anti-spike IgG antibody seropositivity rates to 100% [17].
The result showed a significant difference in anti-spike protein titers based on the vaccination status and the history of COVID-19. Participants who received a full dose of vaccine (4 vaccination steps) with previously SARS-CoV-2-infected resulted in lower anti-spike protein (9,174.40 MFI) compared to those given a full dose of vaccine with no history of infection (11,380.11). This could be related to the documentation of COVID-19 because the asymptomatic participant may not have received any SARS-CoV-2 test and remained undiagnosed. Limitations in diagnostic tools during the early stages of the pandemic, a reluctance to perform testing, or fear of testing during the pandemic peak, and following guidelines that do not necessitate testing in the absence of symptoms might also contribute to bias in documentation, leading to no confirmed cases of COVID-19. This scenario underscores the complexity of interpreting serological data without considering the clinical and epidemiological contexts. In addition, there is evidence that the risk of symptomatic COVID-19 decreased with increasing levels of anti-spike IgG [27].
Natural infection triggers an adaptive immune response to structural proteins, with T cells and neutralizing antibodies targeting the receptor-binding domain (RBD) of the spike protein to prevent virus interaction with the angiotensin converting enzyme (ACE) 2 receptor. These antibodies are crucial for COVID-19 protection and correlate with protective immunity against reinfection [27]. However, a study found that the level of anti-spike with 506 BAU/mL was sufficient to prevent 80% of all symptomatic infections with the Alpha (B.1.1.7) variant [27]. Measuring the avidity/affinity of IgG directed towards viral spike protein gives better information and certainty on the protective potential of immune responses against SARS-CoV-2. This approach specifies the composition of the IgG population that is relevant for protection [28]. Achieving avidity determination of IgG in future serological studies would be needed to broaden the horizon of the protection effect in HCWs.
Elderly individuals tend to have a lower antibody response to vaccinations compared to younger ones. This age-related decrease in neutralization capacity is attributed to the decline in innate and adaptive immunity, with elderly participants showing lower antibody titers due to impaired T cell and plasma cell function [26, 29].
Gender differences in antibody responses have been observed, with females generally showing higher antibody responses to most vaccines than males. Although the result was not significant, other studies also confirmed this difference [30, 31]. The disparity is attributed to the innate and adaptive immune systems of females responding more robustly and quickly to vaccinations. This phenomenon is influenced by hormonal, behavioral, and genetic factors. Androgens may play a role in the gender disparity in disease severity, regulating the transcription of the transmembrane protease, serine 2 (TMPRSS2), essential for SARS-CoV-2 infectivity [29, 32].
Regarding the relationship between vaccination status and IgG antibody titers, significant differences were observed in specific spike protein IgG titers among different vaccine combinations and vaccination statuses, including full dose and infected groups. Previous studies have shown that mixed vaccine regimens elicited stronger and more durable antibody responses than single-vaccine regimens, particularly with mRNA-based vaccines known for inducing the expression of SARS-CoV-2 spike protein, thereby facilitating the development of memory immune cells [17].
Natural infection elicits T cells and neutralizing antibodies, which effectively prevent the virus from interacting with the ACE2 receptor [33]. Interestingly, this study found that participants with complete vaccination and no COVID-19 history had the highest mean antibody levels. In contrast, a previous report stated that unvaccinated individuals who recovered from infection exhibited higher immunity levels [34].
The result showed a robust and durable IgG response among HCWs, influenced by vaccination, combination, and elapsed time since the last dose. The study underscores the critical importance of full vaccination regimens in maintaining immunity and the necessity for continued monitoring to inform potential future booster strategies in the post-pandemic context.
HCWs interfering during the COVID-19 pandemic were found to be very prone to infection. The study sample represented the hospital population from different work units but was not fairly distributed regarding sex (male 40.8% vs. female 59.2%) and age (≤ 40 years old 87.4%, > 40 years old 12.6%). Additionally, other factors possibly related to the immunity status should be considered. For instance, the health status, quality of life, cardio-respiratory performance, and psychological variables could be of great relevance, as stated by Rolando et al. [35] and Osses-Anguita et al. [36].
IgG antibody testing performed did not evaluate the neutralizing capability derived from observations of analysis and T-cell responses. Furthermore, the MFI units used were not converted into antibody units (AU), which may affect the comparability and interpretation of the results in the broader context of immunological study. Further investigations with more representative participants, larger sample size, and the use of control groups in analyzing data could raise the generalizability of the results.
In conclusion, this study showed that HCWs have maintained a strong humoral immune response against SARS-CoV-2, even more than 30 months post-vaccination. This underscored the effectiveness of the vaccination program as well as its critical role in long-term pandemic management and mitigation. The results can assist future booster strategies and guide healthcare policies in managing potential future outbreaks. In addition, continued surveillance and studies in this area are considered essential.
COVID-19: Coronavirus disease 2019
DBS: dried blood spot
HCWs: healthcare workers
IgG: Immunoglobulin G
MFI: mean fluorescence intensity
mRNA: messenger RNA
rpm: rotation per minute
SARS-CoV-2: severe acute respiratory syndrome coronavirus 2
xMAP: multi-antigen profiling
The supplementary materials for this article are available at: https://www.explorationpub.com/uploads/Article/file/1003166_sup_1.pdf.
The authors are grateful to all participants who have consented to include the data for the study as well as Isnaeni, a virology laboratory analyst, for the assistance in processing samples. The authors are also grateful to the First Affiliated Hospital of Dr. Wahidin Sudirohusodo as well as Prihantono, Nurpudji Astuti Taslim, Firdaus Kasim, and Andriany Qanitha for tutoring and assisting in compiling the manuscript.
IN, EA, NAT, ID, BN, SN: Conceptualization, Investigation, Methodology, Data curation, Validation. JH, AA: Statistical analysis, Supervision. IN, RR: Writing—original draft, Writing—review & editing. MAGM: Writing—review & editing. All authors read and approved the submitted version.
The authors declare that there are no conflicts of interest.
All participants gave written informed consent following the Declaration of Helsinki. The Ethical Committee of the Medical Faculty of Hasanuddin University approved the procedure applied in this study (750/UN4.6.4.5.31/PP36/2023).
Informed consent to participate in the study was obtained from all participants.
The consent for publication has been obtained from all participants.
Data are available in the supplementary materials. Supporting raw data for this study are available from the corresponding author upon reasonable inquiry.
Not applicable.
© The Author(s) 2024.
Copyright: © The Author(s) 2024. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
View: 3367
Download: 34
Times Cited: 0
Vijay Mishra ... Yachana Mishra
