Abstract
Immune response, inflammation, and lipid metabolism have important effects on cancer development and progression. Several proteins in tumoral cells and/or tumor microenvironment are involved in any of these processes, whereas some of them participate in all three, such as the zinc finger E-box-binding homeobox 1 (ZEB1) protein. This protein has been proposed to have an important role in invasion and metastasis of cancer cells, as well as to be involved in malignant transformation and resistance to cancer treatments. So, in this study, we present the participation of ZEB1 in immune, inflammatory, and membrane remodeling (lipid metabolism) processes, as well as its interaction with proteins that participate in them. Due to the importance of ZEB1 in cancer progression, it may be a potential biomarker of cancer prognosis and a target for the development of new cancer therapies.
Keywords
ZEB1, cancer, immune response, inflammation, membrane, lipids, epithelial-mesenchymal transition, metastasisIntroduction
The immune response has the dual capacity to promote and suppress tumor growth. Tumor development, progression, and even metastasis are likely to be continuously subject to a balance between immunosurveillance and tumor-promoting chronic inflammation. Cancer cells may express “non-self” antigens as a result of genetic mutations, and these new antigens can be presented to lymphocytes to trigger a response against the tumor. However, tumors have taken advantage of several mechanisms to avoid the immune response, such as immunoediting, the selection of less immunogenic tumor cells, as well as the formation of an immunosuppressive tumoral microenvironment through regulatory cells, downregulation of antigen presentation, suppressive mediators, induction of anergy or tolerance, immune deviation, and apoptosis of immune cells [1, 2].
The association between inflammation and cancer has been established since the 19th century. Chronic inflammation caused by persistent infections, autoimmunity, environment, and dietary exposure, among others; as well as tumor-associated inflammation and therapy-induced inflammation has been related to cancer development and progression [2, 3]. Reactive oxygen and nitrogen species produced during inflammation can cause deoxyribonucleic acid (DNA) damage, which in turn can provoke mutations that initiate and promote cancer [4]. An inflammatory microenvironment in the tumor can also induce angiogenesis, invasion, and metastasis. Moreover, inflammation can affect the response to therapy in a positive or negative manner [2].
Tumor alters lipid metabolism in its favor for its development, survival, and even, invasion and metastasis. Cancer cells have the ability to synthesize de novo (lipogenesis) and exogenously uptake fatty acids, which can be used for energy production through β-oxidation, and storage in lipid droplets as reserves for stress situations. Lipids are also required for cell membrane synthesis during proliferation, and can modify the structure of this, making it more saturated and rigid, which protects against oxidative stress and confers drug resistance, or more flexible, allowing cell migration resulting in metastasis. Moreover, membrane lipids can aid cell surface receptor clustering and downstream oncogenic signaling. In the tumor microenvironment, altered lipid metabolism can stimulate tumor-promoting inflammation, enhance angiogenesis, influence stromal cells, and affect the immune cell compartment, allowing escape from the immune system [5–7].
Immune response, inflammation, and lipid metabolism are involved in several cancer processes, such as development, progression, angiogenesis, invasion and metastasis, and even, drug resistance. Several proteins are related to these processes in tumor cells and the tumor microenvironment, some of them participate in all three processes, such is the case of the zinc finger E-box-binding homeobox 1 (ZEB1) protein. This protein has been proposed to play an important role in cancer cells invasion, migration and metastasis; furthermore, ZEB1 could participate in malignant transformation and the tumor onset, as well as in evasion of the immune response and resistance to cancer treatments [8]. That is why in this short communication we highlight the importance of ZEB1 in the evasion of antitumor immune responses, as well as in inflammation and membrane remodeling processes during neoplasia progression (as shown in Figure 1).
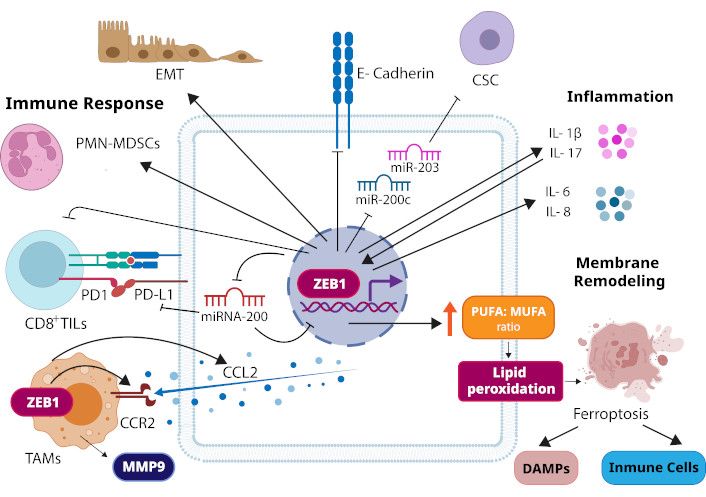
Roles of ZEB1 in neoplasia. ZEB1 induces epithelial-mesenchymal transition (EMT) in carcinoma cells. During EMT, ZEB1 suppresses the expression of epithelial markers such as E-cadherin, a transmembrane protein that constitutes adherens junctions. ZEB1 can also promote the cellular transition to cancer stem cells (CSCs). It can repress the expression of two microRNAs (miRNAs) that inhibit stemness, miR-200c and miR-203. ZEB1 has been related to suppression of antitumoral immune responses. It promotes the recruitment of polymorphonuclear myeloid-derived suppressor cells (PMN-MDSCs) to the tumor site. It also represses miRNA-200, leading to an increase in PD-L1 on tumor cells, which through binding to its receptor PD1 provokes the suppression of CD8+ tumor-infiltrating lymphocytes (CD8+ TILs). Expression of ZEB1 in tumor-associated macrophages (TAMs) induces upregulation of CCR2 (the receptor of chemokine CCL2) and MMP9, as well as, in cancer cells, it induces CCL2, triggering the recruitment of TAMs to the tumor microenvironment. In cancer-related inflammation, pro-inflammatory cytokines IL-1β and IL-17 promote the increase and activation of ZEB1. In turn, ZEB1 induces the production of IL-1β and inflammatory response genes, such as IL-6 and IL-8. ZEB1 increases the PUFA/MUFA ratio of the membrane phospholipids, which may result in peroxidation of PUFAs to trigger ferroptosis. This cell death may cause the release of damage-associated molecular patterns (DAMPs) to trigger innate immune responses and induce cytotoxic T cell responses or may affect the number and functions of immune cells. CCL2: C-C motif chemokine ligand 2; CCR2: C-C motif chemokine receptor 2; IL-1β: interleukin-1β; MMP9: matrix metalloproteinase-9; MUFA: monounsaturated fatty acid; PD1: programmed cell death protein 1; PD-L1: PD1 ligand 1; PUFA: polyunsaturated fatty acid
ZEB1 protein structure and functions
ZEB1 protein, also known as δEF1, Nil-2-a, TCF8, among others, is a member of the ZEB transcription factor family [9]. This protein contains two zinc finger clusters towards its N- and C-terminal ends, which bind to DNA at the E-box and E-box-like sequences (5’-CANNTG-3’) in the regulatory regions of target genes, and a central POU-like homeodomain. ZEB1 also presents several protein binding domains that interact with other transcription factors, as well as with coactivators and corepressors. Therefore, it may act as a transcriptional repressor or activator, depending on the proteins with which it interacts [8–10].
During embryonic development, ZEB1 has a critical role in the activation of epithelial-mesenchymal transition (EMT), a very important process in neural crest development, heart valve development, mesoderm formation, and secondary palate formation, among others. On the other hand, the physiological function of ZEB1 in adults is still largely unknown [11].
In carcinoma cells, ZEB1 participates in the induction of migratory, invasive and metastatic capabilities through the activation of the EMT process (see Figure 1). In addition, ZEB1 may have a role in the early steps of tumorigenesis, including malignant transformation and tumor initiation, due to that this protein is able to prevent senescence and apoptosis, as well as to promote the G1-S cell-cycle progression. ZEB1 has also been proposed to contribute to tumor progression; regulate stemness, inducing the transition of non-cancer stem cells (CSCs) to CSCs; and confer resistance to chemotherapy, radiotherapy, and new anticancer therapies, such as targeted therapies and immunotherapies [8]. High ZEB1 expression has implications for poor prognosis for both progression and treatment [12].
According to Dongre and Weinberg [13], “EMT is a reversible cellular programme that transiently places epithelial cells into quasi-mesenchymal cell states”. During this process, epithelial cells lose their cell-cell and cell-basement membrane junctions, as well as their apical-basal cell polarity and cobblestone appearance, to gradually acquire a spindle-shaped mesenchymal morphology with front-to-back polarity, as well as motile and invasive capabilities [13]. In addition to its crucial role in embryogenesis, EMT is also important for wound healing, tissue fibrosis, and even cancer progression, particularly in invasion and metastasis, as well as therapeutic resistance and the acquisition of a CSC state [13–15].
The EMT process is activated by several signaling molecules of the tumor microenvironment. These molecules stimulate various signaling pathways, which in turn activate a set of EMT-inducing transcription factors, among which is ZEB1. These transcription factors are responsible for suppressing the expression of epithelial markers such as E-cadherin, occludin, claudin, mucin-1, among others; as well as activating mesenchymal markers such as N-cadherin, vimentin, matrix metalloproteases, and others [15].
Several microRNAs (miRNAs) can downregulate the expression of ZEB1 directly or indirectly, thereby inhibiting its effects, such as EMT induction, migration, invasion, and metastasis, as well as anticancer treatment resistance. In turn, ZEB1 can repress these miRNAs to promote tumorigenesis, thus establishing a negative feedback loop [16, 17] (as shown in Figure 1). As example of this, it has been reported that ZEB1 directly represses the expression of miRNA-200 family members, miR-141 and miR-200c, which activate epithelial differentiation, and ZEB1 is a downregulated target of these miRNAs, regulating in this manner EMT [18]. In addition, ZEB1 can directly repress miR-203 expression, which together with miR-200c can inhibit stemness. Thus, ZEB1 could link EMT activation and CSCs [19]. On the other hand, long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs) can modulate the miRNAs/ZEB1 axes. These can act as competitive endogenous RNAs (ceRNAs) to capture miRNAs responsible for ZEB1 downregulation, allowing increases in ZEB1 expression and its consequences on tumorigenesis [16].
ZEB1 and immune response
ZEB1 has been related to suppression of antitumoral immune responses. Its expression in breast cancer cells promotes the accumulation of polymorphonuclear myeloid-derived suppressor cells (PMN-MDSCs) at tumor site [20]. Moreover, it has been proposed that ZEB1 expression is inversely correlated with immune cell abundance in breast, colorectal, lung, prostate, and pancreatic adenocarcinoma. In breast cancer, ZEB1 and its transcriptional expression signature are associated with reduced immune activity and worse overall survival [21]. In melanoma, ZEB1 expression is associated with decreased CD8+ T cell recruitment, provoking tumor immune evasion. This protein represses the secretion of T cell-attracting chemokines, such as CXCL10 [22].
Cytotoxic CD8+ tumor-infiltrating lymphocytes (CD8+ TILs) can kill cancer cells and have been associated with good prognosis in several types of cancer [23]. However, CD8+ TILs can suffer exhaustion by prolonged exposure to cancer cells, through the interactions of the co-inhibitory receptor programmed cell death protein 1 (PD1) on CD8+ TILs surface with its ligand PD1 ligand 1 (PD-L1) expressed on cancer cells and other cell types in the tumor stroma. It has been shown that PD-L1 is directly downregulated by the miRNA-200 family members, by binding to two sites in the 3’-untranslated region (UTR) of PD-L1. Moreover, ZEB1 has been suggested to repress miRNA-200, leading to increased PD-L1 on tumor cells, provoking suppression of CD8+ TILs and facilitating metastasis [24] (Figure 1).
Tumor-associated macrophages (TAMs) are an important component of the tumor microenvironment. They have been related to tumor growth, metastasis and therapy failure [25]. Expression of ZEB1 in TAMs induces a tumor-promoting phenotype, upregulation of CCR2 (the receptor of chemokine CCL2) and MMP9, a matrix metalloproteinase. On the other hand, in cancer cells, ZEB1 expression by TAMs induces CCL2 (a chemoattractant of monocytes), CD74 (a transcription factor that activates CCL2 and survival genes expression), and a mesenchymal/stem-like phenotype. In ovarian cancer, ZEB1 expression in TAMs and ZEB1/CCL2/CD74 high expression in cancer cells predict poorer survival [26].
ZEB1 and inflammation
ZEB1 has been related to the inflammatory process in cancer development in several studies, here we mention some of these (see Figure 1). Interleukin-1β (IL-1β) is a pro-inflammatory cytokine implicated in cancer-related inflammation. This can be produced by immune, stromal, and tumor cells, and its levels are increased in various cancers. It has been demonstrated that IL-1β may promote CSCs self-renewal and EMT through ZEB1 activation in colon cancer cells, which may contribute to tumoral progression [27]. In head and neck squamous cell carcinoma (HNSCC) cells, IL-1β provokes an increase in ZEB1 and a decrease in E-cadherin at the mRNA level, as well as enhances the binding of ZEB1 to the E-cadherin promoter at chromatin level [28]. IL-17 is another inflammatory cytokine that is also involved in cancer progression. This cytokine induces EMT via upregulation and activation of ZEB1 in lung cancer cells, and nuclear factor-κB (NF-κB) may participate in these processes to promote cell migration and invasion [29]. However, NF-κB has also been related to tumor suppression in tissue cells and immune cells in the tumor microenvironment, indicating that its roles in cancer development may be context-specific [30, 31]. On the other hand, in breast cancer cells, ZEB1 induces the expression of inflammatory response genes, especially of IL-6 and IL-8, two cytokines whose elevated expression has been correlated with poor survival. In addition, ZEB1 increases the proliferation and tumor growth of breast cancer cells [20].
ZEB1 has been proposed to be an important driver of intestinal inflammation (colitis) and inflammatory colorectal cancer (CRC). Its expression is absent in epithelial cells of healthy colon, but is induced during an intestinal inflammatory condition, such as colitis. In turn, ZEB1 promotes intestinal inflammation and inflammatory-induced CRC tumorigenesis, and does this, at least in part, inhibiting the expression of N-methylpurine DNA glycosylase (MPG), an enzyme implicated in DNA base excision repair. In addition, the expression of ZEB1 in CRC cells induces the production of reactive oxygen species (ROS) and IL-1β by macrophages. This could stimulate a positive loop to exacerbate inflammation, oxidative stress, and DNA damage [32].
ROS have a dual role in cancer. Excess of ROS can promote tumor initiation, progression, invasion, and metastasis, through DNA mutations, inhibition of T cells and natural killer (NK) cells, recruitment and M2 polarization of macrophages, as well as induction of EMT in tumor cells. On the other hand, massive accumulation of ROS can suppress tumor progression by inhibiting tumor cell proliferation and inducing cancer cells death via apoptosis and ferroptosis. However, cancer cells activate mechanisms to counteract these effects [33]. In addition, ROS have been identified as a main cause of EMT in cancer cells. Elevated ROS levels can induce the activation of NF-κB signaling, leading to the upregulation of ZEB1. Although it has been suggested that ROS may also inhibit NF-κB signaling [33–35].
Employing an in vivo model of metastatic latency of mammary carcinoma cells in the lung parenchyma, ZEB1 has been demonstrated to awaken dormant cells to produce metastases, and to generate metastasis-initiating cells. In addition, ZEB1 is important in the awakening of latent cells induced by inflammation caused by lipopolysaccharide treatment [36].
ZEB1 and membrane remodeling
ZEB1 has an impact on different metabolic pathways that relate to the microenvironment of the cell membrane. For example, the expression of E-cadherin in EMT affects the tumor microenvironment and the response of immune cells. Likewise, ZEB1 also has an impact on the MAPK pathways. In addition, at membrane level, ZEB1 participates in the repression of basal membrane synthesis, promoting remodeling of the extracellular matrix and tumor cell invasion [12, 37].
The cell death pathway known as ferroptosis, present in the EMT process, is driven by the peroxidation of polyunsaturated fatty acids (PUFAs). ZEB1 enriches PUFAs in phospholipids and enhances peroxidation by increasing the PUFA/MUFA (monounsaturated fatty acid) ratio of the membrane by regulating the expression of enzymes that control the ratio of unsaturated to saturated fatty acids. These favor membrane fluidity and contribute to bilayer remodeling during the EMT [8, 38]. Ferroptosis of cancer cells may cause the release of damage-associated molecular patterns (DAMPs) which may bind to pattern recognition receptors (PRRs) to trigger innate immune responses and may be presented to induce cytotoxic T cell responses. On the other hand, ferroptosis may affect the number and functions of the immune cells themselves (Figure 1). Therefore, it is unclear whether ferroptosis has an anti- or pro-tumoral effect, and it might be context dependent [39].
Membrane remodeling resulting from variation in ZEB1 expression may favor the pathogenesis of neoplasia, since it affects diverse metabolic pathways that affect the immune and inflammatory response and that have been studied in systems other than neoplasia, such as the study in eosinophilic chronic rhinosinusitis where the contribution of ALOX15+ M2 macrophages is evident, or the remodeling of chromatin in inflammation and fibrosis where ZEB1 silences the expression of the interferon regulatory factor 1 (IRF1), where this same behavior is related to inflammation and cancer processes [40, 41].
The effect that ZEB1 has on lipid changes in the membrane and its contribution to the process of metastasis and malignancy of cancer cells, place it as a possible therapeutic target or biomarker as it is present in the processes of immune response, inflammation, and membrane remodeling, which could contribute to the paradigm of the influence that the dynamics and organization of all membrane components have during neoplasia [42]. Singler and Nicolson [43] 1972 demonstrated that the membrane is a fluid mosaic consisting of a heterogeneous arrangement of lipids and proteins. The fluid mosaic model has generated various studies on the importance of membrane lipids, such as the formation of microdomains known as lipid rafts and their participation in neoplasia [44], or the importance of membrane remodeling through the increase in the concentration of sPLA2 during signaling in inflammatory processes, tissue remodeling or carcinogenic processes, for example, where the formation of domains and the effect they have on membrane fluidity has been demonstrated [45]. Therefore, changes in protein function or signaling processes may be attributed to the change in membrane fluidity during the generation of lipid microdomains, when the heterogeneous microenvironment surrounding membrane proteins is modified, which would possibly contribute to the disruptive effect that favors the EMT process during tumor formation [38, 46], as exemplified in Figure 2.
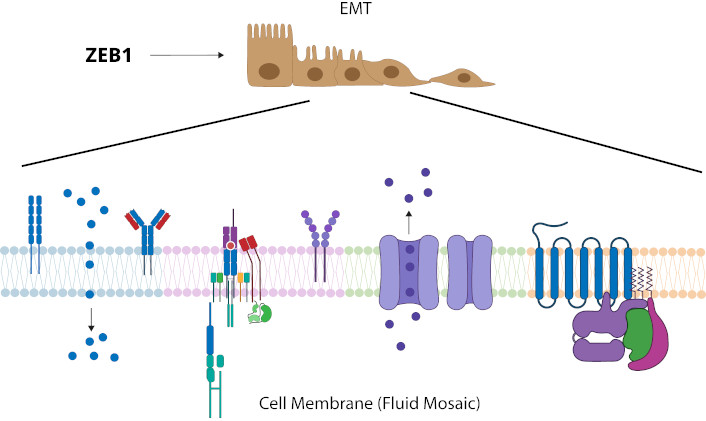
Change of membrane remodeling and fluidity (PUFA/MUFA) from role of ZEB1 in neoplasia. ZEB1 has been shown to induce EMT in neoplastic processes, and there is a change in the PUFA/MUFA ratio that favors the modification of membrane fluidity and membrane remodeling based on the formation of microdomains. Changes in the membrane lipid-protein microenvironment may impact on the different functions of membrane proteins or signaling. EMT: epithelial-mesenchymal transition; MUFA: monounsaturated fatty acid; PUFA: polyunsaturated fatty acid
ZEB1 related proteins
A bibliographic search was carried out through CONNECTED PAPERS (https://www.connectedpapers.com/) and PubMed [National Center for Biotechnology Information (NCBI), https://pubmed.ncbi.nlm.nih.gov/] for possible interactions of ZEB1 with proteins involved in immune response, inflammation or membrane remodeling (looking for a change in membrane lipids for membrane remodeling). Metabolic pathways were consulted on Roche Biochemical Pathways (https://www.roche.com/about/philanthropy/science-education/biochemical-pathways) and related articles. As a result of this search, 25 proteins correlated with ZEB1 variations during a neoplastic process were identified. The distribution of the proteins was grouped according to their participation in the processes of immune response, inflammation and membrane remodeling (lipids changes), as presented in the Venn diagram described in Figure 3.
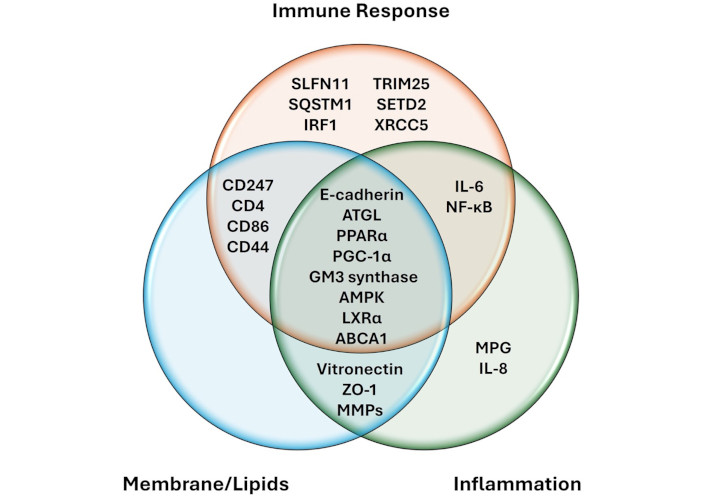
Relationship of ZEB1 with proteins involved in immune response, inflammation and membrane remodeling. ZEB1 interacts directly or indirectly with other proteins related to immune response, inflamation and changes in membrane lipids, some of these proteins participate in two or three of these processes, as indicated in the Venn diagram. AMPK: AMP-activated protein kinase; ATGL: adipose triglyceride lipase; IL-6: interleukin-6; IRF1: interferon regulatory factor 1; LXRα: liver X receptor alpha; MMPs: matrix metalloproteinases; MPG: N-methylpurine DNA glycosylase; NF-κB: nuclear factor-κB; PGC-1α: peroxisome proliferator-activated receptor gamma coactivator 1-alpha; PPARα: peroxisome proliferator-activated receptor alpha; SETD2: SET domain containing 2; SQSTM1: sequestosome-1; SLFN11: schlafen family member 11; TRIM25: tripartite motif containing 25; XRCC5: X-ray repair cross-complementing protein 5; ZO-1: zonula occludens protein 1
ZEB1 regulation participates in intercellular and intracellular signaling processes modulated by different protein signaling pathways, as well as methylation at the genetic and epigenetic level. It has been studied that the cellular response to ZEB1 regulation has implications of EMT in carcinogenic processes, because EMT promotes chemoresistance, invasion and metastasis, and generates circulating tumor cells (CTC), CSCs and aggressive cancer cells.
In intercellular processes, EMT modulation is induced by the positive or negative regulation of ZEB1, through the suppression of, for example, IRF1 that influences the overexpression of MTMR2 [47], or the regulation of ZEB1 by TRIM25, which promotes oncogenic activities in breast cancer [48]. On the other hand, in extracellular processes, the expression of ZEB1 also has an implication through processes of ROS and IL-1β generation in macrophages that induce an inhibitory response of MPG in CRC cells [32].
At the methylation level, the regulation of SETD2 through histone methylation can be combined with the function of ZEB1 during the promotion of tumor progression through the EMT transition [49]. Another study describes that downregulation of the transcription factor ZEB1 upregulates E-cadherin and TROP2, thus sensitizing to immunotherapeutic treatments. Because TROP2 and SLFN11 expression can be modulated epigenetically and, therefore when combined with a demethylating agent to act as a therapy for tumors with low TROP2 or SLFN11 expression [50].
In breast cancer caused by mutant RAS, it has been shown that inhibition of autophagy induces the EMT transition, which is associated with the participation of ZEB1 and the activation of the NF-κB pathway through SQSTM1/p62, so the deletion of SQSTM1 eliminates the activation of the NF-κB pathway, favoring the blocking of EMT induction, this finding opens the possibility of considering this pathway for future treatments [51–53].
A study of cancer cells and their cell death response exerted by iron and oxygen radical-mediated peroxidation of phospholipids containing polyunsaturated fatty acids, in an EMT process favored by ZEB1 and stimulated by TGF-β, suggests a combination of ferroptosis activators and SCD inhibitors for the treatment of aggressive cancers that express ZEB1 in high amounts [38].
As can be seen, the implications that ZEB1 modulation has on expression, methylation and signaling in the carcinogenic process and its close relationship with EMT induction, makes it a potential biomarker for cancer [54].
Given the participation of ZEB1 in EMT, immune response, inflammation, and membrane remodeling processes during tumor progression, it is an attractive target for the development of new cancer therapies, alone or in combination with other existing ones. Downregulation of ZEB1 expression using miRNAs or small interfering RNAs (siRNAs) can enhance sensitivity of cancer cells to chemotherapy [55, 56] and radiotherapy [11], but more precise and efficient delivery systems are necessary, such as liposomes and nanocarriers. Furthermore, ZEB1 expression can also be reduced indirectly through regulation of upstream targets, such as inhibition of lncRNAs or circRNAs that capture miRNAs involved in ZEB1 downregulation [57], or inhibition of proteins that enhance ZEB1 transcription [58].
Other ZEB1-related proteins that were classified in the Venn diagram are described in Table 1, which indicates the gene and location of the protein, its characteristics and functions, as well as its participation in immune response, inflammation, and membrane remodeling or lipid metabolism. The Table 1 condenses information on important aspects that could help during the design and development of systems aimed at prevention, diagnosis, therapy, or regeneration, focusing the design on a target protein, on the regulatory function of ZEB1 or at the genetic-epigenetic level associated with ZEB1.
Proteins related to ZEB1 involved in immune, inflammatory, and membrane/liposomal changes
| Protein [59–61] | Features and functions | Immune response | Inflammation | Membrane/Lipids |
|---|---|---|---|---|
| 1. E-cadherin(Cadherin-1, epithelial cadherin)GeneCDH1 (CDHE, UVO)LocalizationCell junction, cell membrane, endosome, Golgi apparatus | Transmembrane calcium-dependent cell adhesion glycoprotein that constitutes the adherens junctions. Interacts with cytoplasmic actin through alpha, beta, and gamma catenin [62, 63].CDH1 mutation is associated with cancer [64].Controls adhesion and locomotion in tumors [63]. | Contributes to the maintaining of epithelial barriers which protect the internal tissues.May take part in modulating transepithelial transfer of immune cells.Participates in regulating immune cells in the tumor microenvironment, like dendritic cells (DCs) and macrophages [65]. | Its overexpression suppresses inflammation by influencing inflammatory signaling pathways [66]. | The composition of membrane lipids maintains adherens junctions by affecting junctional proteins such as E-cadherin [67]. |
| 2. IL-6(Interleukin-6)GeneIL6 (BSF2, CDF, HGF, HSF, IFNB2, IL-6)LocalizationSecreted | Cytokine binds to the IL-6 receptor (IL-6R) and this complex associates with gp130, inducing its dimerization to initiate signaling [68, 69].Has a pleiotropic effect on inflammation, immune response, and hematopoiesis. Can induce angiogenesis [70]. | Promotes differentiation of activated B cells into Ig-producing cells.Is indispensable for T helper 17 cells differentiation and inhibits regulatory T cells differentiation.Can promote T follicular helper cells differentiation and production of IL-21 to regulate Ig synthesis [70]. | Presents pro- and anti-inflammatory activities [69].Induces synthesis of acute phase proteins. Activates vascular endothelial cells and induces vascular permeability [70]. | - |
| 3. NF-κB(Nuclear factor-κB: RELA/p65, RELB, NFKB1/p50, REL and NFKB2/p52)GeneRELA, RELB, NFKB1, REL and NFKB2LocalizationNucleus, cytoplasm | Family of transcription factors that regulate innate and adaptive immunity, inflammation, cellular proliferation, differentiation, and survival [71].In cancer, it may have a dual role, depending on the context, it can promote or repress tumor progression [30, 72].It stimulates EMT through the upregulation of inducers like ZEB1 [34]. | Activated in several immune cells that constitute the tumor microenvironment, promoting or repressing tumor development, such as TAMs, MDSCs, DCs, NK cells, and T and B lymphocytes [72]. | Plays an important role in coordinating the entire inflammatory response.Establishes a network with cytokines, like tumor necrosis factor (TNF), IL-1, IL-6, and IL-17A, as well as induces many chemokines, which can promote tumorigenesis and metastasis [72]. | - |
| 4. ATGL(Patatin-like phospholipase domain-containing protein 2, adipose triglyceride lipase)GenePNPLA2 (ATGL)LocalizationLipid droplet, cell membrane, cytoplasm | Enzyme that catalyzes the first reaction of lipolysis, hydrolyzing triacylglycerols to diacylglycerols, in lipid droplets of adipocytes and non-adipocytes [73].Its expression is reduced in different cancers [74]. | Involved in the production of fatty acids by sPLA2, that intervene in signaling pathways of metabolism, inflammation, immunity, and cancer [73]. | Provides an important source of fatty acids such as energy substrates, signaling and precursors of membrane lipids [73]. | |
| 5. PPARα(Peroxisome proliferator-activated receptor alpha)GenePPARA (NR1C1)LocalizationNucleus, cytoplasm | Transcription factor regulated by free fatty acids and a key regulator of lipid metabolism [75, 76].Linked to cancer progression [75]. | Participates in the development, differentiation, and functions of immune cells, such as macrophages and T cells [77]. | One of the key proteins involved in inflammation processes [76]. | A key regulator for the expression and metabolism of lipids [78]. |
| 6. PGC-1α(Peroxisome proliferator-activated receptor gamma coactivator 1-alpha)GenePPARGC1A (PGC1A)LocalizationNucleus | Transcriptional coactivator of steroids and nuclear receptors [76].It has an important role in cancer metabolism. Presents pro- and anti-neoplastic functions [79]. | In CD8+ T cells, it promotes cell activation, memory cells, and antitumor immunity [80, 81].In NK cells, it is important for the effector functions in the response to infection and the control of tumor growth [82]. | Low levels in inflamed tissues increase ROS production and oxidative damage [83]. | It can bind to PPARα, PPARβ/δ, and PPARγ, contributing to the transport and utilization of fatty acids [84]. |
| 7. GM3 synthase(Lactosylceramide alpha 2,3-sialyltransferase)GeneST3GAL5 (SIAT9)LocalizationGolgi apparatus membrane | Enzyme involved in the synthesis of gangliosides, like GM3 [85].GM3 regulates cell adhesion, growth, and motility through alteration of membrane microdomains and the activation of signaling molecules involved in cancer [86]. | Upregulated in CD4+ T cells, in which induces the expression of a-series gangliosides, and downregulated in CD8+ T cells, which express o-series gangliosides.a-series gangliosides are necessary for CD4+ T cells activation, and o-series gangliosides are required for CD8+ T cells activation [87]. | Its participation in inflammation has been studied in pathologies such as renal fibrosis. Its involvement in neoplasia elucidates that it may have an important role in cancer inflammation [88, 89]. | Participates in the organization of glycolipid microdomains of the membrane during cancer [86, 90]. |
| 8. AMPK(AMP-activated protein kinase)GenePRKAA1, PRKAA2, PRKAB1, PRKAB2, PRKAG1, PRKAG2, and PRKAG3LocalizationCytoplasm, nucleus | Protein present in the regulation of cellular energy metabolism [91].It can act as a tumor suppressor by regulating energy levels in cancer [92]. | May regulate activities and functions of immune cells, like macrophages, T cells, B cells, and NK cells, in the tumor microenvironment [93]. | Participates in inflammation signaling by inhibiting IL-1β expression and NF-κB activation [92]. | Participates in lipidic remodeling pathways through ferroptosis [94]. |
| 9. LXRα(Liver X receptor alpha)GeneNR1H3 (LXRA)LocalizationNucleus, cytoplasm | Belongs to the nuclear liver X receptors, which respond to changes in levels of lipid ligands and regulate the transcription of genes involved in lipid metabolism [95]. | In cancer, progranulin expression in macrophages is necessary for efficient efferocytosis by controlling lysosomal acidification via cystic fibrosis transmembrane conductance regulator and the degradation of lysosomal cargo, resulting in LXRα/RXRα-mediated macrophage conversion and upregulation of arginase 1 [96]. | Necessary for the synthesis of cholesterol in response to inflammatory signals.Inhibit proinflammatory genes [95, 97]. | Regulate the expression of genes encoding proteins involved in the absorption, transport, efflux, excretion, and conversion of cholesterol into bile acids.Regulate the metabolism of fatty acids and phospholipids [95]. |
| 10. ABCA1(Phospholipid-transporting ATPase ABCA1)GeneABCA1 (ABC1, CERP)LocalizationCell membrane, endosome | Membrane protein responsible for phospholipids and cholesterol efflux from the cytoplasm [98, 99].Indirectly represses proliferation, invasion, and metastasis of cancer cells by downregulating the level of intracellular cholesterol [100]. | Participates in the activation of anti-tumor Vγ9Vδ2 T lymphocytes, by phosphoantigens, like isopentenyl pyrophosphate (IPP) [101–103]. | Its deficiency is associated with proinflammatory status, with increases in TNF-α, CCL2, and IL-6 levels [104]. | Mediates the transfer of phospholipids and cholesterol into apolipoprotein A-I (apoA-I) to generate nascent high-density lipoprotein (nHDL) particles [98, 99]. |
MDSCs: myeloid-derived suppressor cells; NK: natural killer; ROS: reactive oxygen species; sPLA2: secreted phospholipase A2; TAMs: tumor-associated macrophages
Conclusions
ZEB1 protein has an important role in cancer cells invasion and metastasis by activating EMT, as well as participates in cancer onset, and therapy resistance. This study makes evident the participation of ZEB1 in immune response, inflammation, and membrane remodeling in several types of cancer, as well as its relationship with other proteins that are also involved in these processes. Immunity, inflammation, and membrane altering are of utmost importance in the development and progression of cancer, so ZEB1 may be considered a biomarker of cancer prognosis and a potential target for the development of new preventive or corrective treatments, individually or in combination with existing ones, such as immunotherapy, or through the possible reactivation of the immune response using the interconnected ZEB1 pathways. For this reason, future research with these objectives is necessary.
Abbreviations
| CCL2: | C-C motif chemokine ligand 2 |
| CCR2: | C-C motif chemokine receptor 2 |
| CD8+ TILs: | cytotoxic CD8+ tumor-infiltrating lymphocytes |
| circRNAs: | circular RNAs |
| CRC: | colorectal cancer |
| CSCs: | cancer stem cells |
| EMT: | epithelial-mesenchymal transition |
| IL-1β: | interleukin-1β |
| IRF1: | interferon regulatory factor 1 |
| lncRNAs: | long non-coding RNAs |
| miRNAs: | microRNAs |
| MPG: | N-methylpurine DNA glycosylase |
| MUFA: | monounsaturated fatty acid |
| NF-κB: | nuclear factor-κB |
| NK: | natural killer |
| PD1: | programmed cell death protein 1 |
| PD-L1: | programmed death-ligand 1 |
| PUFAs: | polyunsaturated fatty acids |
| ROS: | reactive oxygen species |
| TAMs: | tumor-associated macrophages |
| ZEB1: | zinc finger E-box-binding homeobox 1 |
Declarations
Acknowledgments
Figures 1 and 2 are generated in the Adobe Illustrator program.
Author contributions
AMRE: Investigation, Writing—review & editing. RRR: Writing—original draft, Investigation. AAVC: Conceptualization, Investigation, Writing—review & editing, Supervision. All authors read and approved the submitted version.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent to publication
Not applicable.
Availability of data and materials
Not applicable.
Funding
We thank CONAHCYT for the project: CONAHCYT Paradigms of Science 2022-320792; the Postdoctoral Fellowship by Mexico; and the Summer Dolphin Program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Copyright
© The Author(s) 2025.
Publisher’s note
Open Exploration maintains a neutral stance on jurisdictional claims in published institutional affiliations and maps. All opinions expressed in this article are the personal views of the author(s) and do not represent the stance of the editorial team or the publisher.