Affiliation:
1Faculty of Medicine, Universiti Sultan Zainal Abidin (UniSZA), Kuala Terengganu 20400, Malaysia
ORCID: https://orcid.org/0000-0001-5219-7317
Affiliation:
1Faculty of Medicine, Universiti Sultan Zainal Abidin (UniSZA), Kuala Terengganu 20400, Malaysia
2Centre for Research in Infectious Diseases and Biotechnology (CeRIDB), Faculty of Medicine, Universiti Sultan Zainal Abidin (UniSZA), Kuala Terengganu 20400, Malaysia
ORCID: https://orcid.org/0000-0002-9644-6925
Affiliation:
1Faculty of Medicine, Universiti Sultan Zainal Abidin (UniSZA), Kuala Terengganu 20400, Malaysia
ORCID: https://orcid.org/0000-0002-4091-0610
Affiliation:
1Faculty of Medicine, Universiti Sultan Zainal Abidin (UniSZA), Kuala Terengganu 20400, Malaysia
Email: malikamonov@unisza.edu.my
ORCID: https://orcid.org/0000-0001-8279-8471
Explor Med. 2024;5:626–640 DOI: https://doi.org/10.37349/emed.2024.00245
Received: May 14, 2024 Accepted: August 30, 2024 Published: October 13, 2024
Academic Editor: Lee M. Wetzler, Boston University School of Medicine, USA
Streptococcus pneumoniae (S. pneumoniae), which is a Gram-positive diplococcus, has emerged as a significant human pathogen. It is a primary cause of bacterial pneumonia, otitis media, meningitis, and septicemia, leading to a considerable impact on global morbidity and mortality. The investigation of S. pneumoniae and its virulence factors has resulted in the identification of surface endonuclease A (EndA). EndA functions in DNA uptake during natural transformation and plays a significant role in gene transfer. The ability of S. pneumoniae to degrade neutrophil extracellular traps (NETs) enhances its virulence and invasive potential in pneumococcal infections. NETosis occurs when neutrophils release chromatin into the extracellular space to form NETs, capturing and neutralizing pathogens. Currently, NETosis can be induced by several microbes, particulate matter, and sterile stimuli through distinct cellular mechanisms, and this includes the involvement of EndA in S. pneumoniae. Here, we reviewed the cellular functions of EndA, its role in S. pneumoniae as a virulence factor in relation to NETosis, its relationship to immunogenicity, and its involvement in several diseases. The discovery of this relationship would significantly impact therapeutic technology in reducing disease burden, especially pneumococcal infections.
The incidence and mortality due to Streptococcus pneumoniae (pneumococcus) infections are increasing primarily due to increasing antimicrobial resistance and the development of novel virulence mechanisms causing threats to global health and incurring high economic costs to society [1, 2]. The bacterium is a highly adaptive commensal that commonly colonizes the upper respiratory tract of healthy individuals; however, colonized individuals serve as a practical reservoir for S. pneumoniae, facilitating the transmission of the bacteria in the community. The pneumococcus is a leading cause of non-invasive infections such as otitis media and sinusitis, but it can also spread to sterile tissues and organs, leading to invasive diseases such as pneumonia and meningitis, which cause significant morbidity and mortality [3].
Traditionally, pneumococcal diseases were treated with antibiotics and prevented with polysaccharide-based and pneumococcal conjugate vaccines. The introduction of vaccines has resulted in serotype replacement, whereby nonvaccine serotypes have become more prevalent among both asymptomatic carriers and symptomatic cases, indicating the rapid adaptation of the pathogen to the selective pressure exerted by vaccines. Still, S. pneumoniae remains the leading cause of community-acquired pneumonia, otitis media, and meningitis [4]. Unfortunately, the threat of pneumococcal disease remains high due to the dramatic increase in antibiotic resistance [5]. The case-fatality ratio of pneumococcal meningitis in 2015 was around 25–27% in Europe and the Americas, but in the same year in Africa, it was 61% [5, 6].
Various virulence factors in S. pneumoniae, including capsule, pneumolysin, pneumococcal surface adhesin A, and pneumococcal surface protein A, have been found to play crucial roles in mediating adherence, invasion, and evading the host’s immune system [7, 8]. These virulence factors give S. pneumoniae the capacity to escape the host’s immune defense, provide resistance to antimicrobial agents, and establish infection in the respiratory tract. Among the virulence factors, endonuclease A (EndA) offers great attention in S. pneumoniae due to its role in facilitating the escape of the bacterium from the host immune response. EndA assists the bacterium in evading neutrophil phagocytosis by breaking down neutrophil extracellular traps (NETs), and its nuclease activity is required for bacterial transformation to acquire novel characteristics, including resistance to antibiotics [9]. The role of the surface endonuclease in DNA uptake during transformation is important for S. pneumoniae to establish invasive pneumococcal infections by facilitating gene transfer and promoting genetic diversity. Hence, understanding the properties, mechanisms, and specificities of EndA, primarily as a virulence factor in S. pneumoniae, could provide novel ideas for preventing the spread and invasion of the bacterium to cause severe disease.
The success of bacteria in upholding their defense mechanisms against invading genomes is closely tied to the presence of methylases, a crucial component of the restriction-modification (R-M) system within a bacterial species. There are two opposite enzymatic activities involved in the R-M systems: methyltransferase (MTase) and restriction endonuclease (REase). While MTase activity ensures that self and non-self DNA are distinguished from one another, REase recognizes explicitly and cleaves foreign DNA sequences at specific sequences. The successful discriminatory mechanism of the MTase enzyme is achieved by transferring methyl groups to the same specific DNA sequence recognized by the REase within the host’s genome [10, 11].
Numerous investigations have demonstrated that the products produced by foreign DNA restriction by REases have the potential to promote homologous recombination with the host genome [12, 13]. Two potential biological functions of homologous recombination stimulation have been identified and proposed [14]: (i) preventing unintentional R-M-mediated mortality in the host and (ii) generating genetic variety by promoting recombination across related species. It was also discovered that restriction due to REases also contributes to nonhomologous recombination. Many foreign DNA (that lack homology) could be recombined and integrated into recipient genomes using a slight stretch of homologous DNA sequences at one end [15]. Furthermore, data suggests that R-M systems may introduce genomic rearrangements [16, 17]. Consequently, it is conceivable that R-M systems contribute to genomic diversity by promoting homologous recombination and participating in nonhomologous genome rearrangements.
Generally, all these molecular mechanisms contribute to genome plasticity in prokaryotes, along with HGT [18], represent a significant source of novel genetic information in prokaryotes [19, 20] and thus, act as a dominant force in the evolution of bacteria [21]. Griffith initially discovered natural transformation in S. pneumoniae nearly a century ago [22]. Over the past 90 years, S. pneumoniae has been a paradigm for this crucial HGT phenomenon. Avery et al. [23] conducted extensive studies on this discovery, affirming that DNA is the hereditary material transferred to transformed S. pneumoniae cells. Gram-positive and Gram-negative bacteria can exchange genetic material through bacterial transformation, which entails taking in and integrating foreign DNA into the recipient genome [24, 25]. Since then, S. pneumoniae has been used as a model organism to help understand the molecular mechanisms of natural transformation.
S. pneumoniae undergoes four distinct steps in its transformation process: (i) the bacterial cell must monitor external and internal signals to decide when to turn on the competent state; (ii) early and late competence proteins must be expressed to allow for the development of natural competence; (iii) competent pneumococci secrete a murein hydrolase that lyses susceptible neighboring cells to capture homologous DNA from other species of streptococcal species that share the same niche or other pneumococci; and finally, (iv) transcription of the competence genes is stopped to end the competence state [26]. The genetic flexibility of S. pneumoniae is determined by this spontaneous transformation mechanism as has been demonstrated by multiple investigations including Chewapreecha et al. [27], Croucher et al. [28], and Engelmoer et al. [29].
The inherent habitat of S. pneumoniae consists of multispecies biofilms located in the human nasopharynx [30]. Alterations in the particular environment prompt stress responses that may initiate competence development in S. pneumoniae. Nevertheless, for the recipient strain to acquire novel advantageous characteristics that facilitate the survival of stressed pneumococci under unfavorable circumstances, it must be genetically distinct from the donor strain. Consequently, mechanisms exist for capable pneumococci to access homologous DNA in a setting where unrelated DNA prevails. One potential scenario is the aggregation of different pneumococcal strains into microcolonies within a specific habitat, forming a cohesive microbial community, thereby establishing a uniform environment conducive to gene transfer [26]. Besides that, the transformation process in S. pneumoniae is multifaceted, involving various vital components, namely ComEA, EndA, ComEC, and ComFA proteins [31]. Transformations occur at the mid-cell, where EndA is selectively recruited to this region during the competence phase, indicating the crucial role of EndA in the DNA uptake complex and natural transformation of S. pneumoniae [31], as shown in Figure 1.
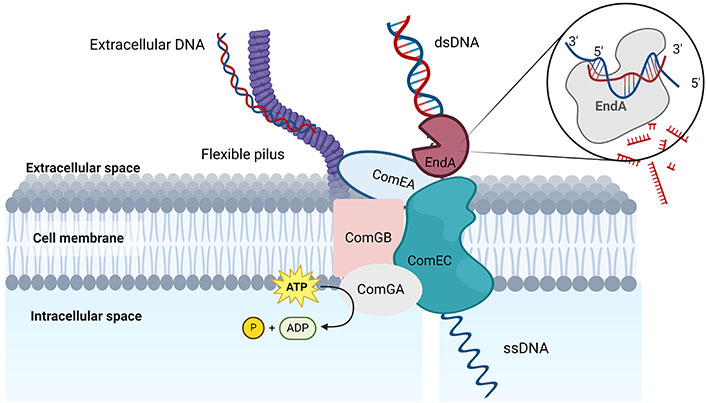
The model illustrates the essential elements of the DNA uptake system in S. pneumoniae. On the surface of bacteria, an extended type IV-like transformation pilus is constructed, which serves the purpose of directly adhering to exogenous DNA. The DNA that has been captured is transported across the cell wall by an unidentified ATPase enzyme to enable the DNA to interact with the DNA receptor ComEA and the transmembrane channel protein ComEC. In an alternative manner, the flexible pilus facilitates the direct transport of captured DNA to cell surface receptors without the involvement of a retraction ATPase. dsDNA is cleaved by the EndA nuclease, leading to the entry of ssDNA into the cytoplasm via the ComEC pore [31]. dsDNA: double-stranded DNA; EndA: endonuclease A; ssDNA: single-stranded DNA. Created in BioRender. Bio, U. (2024) BioRender.com/q92h591
Kohoutová [32] first proposed EndA as playing a significant role in transformation, and this was subsequently confirmed by other researchers [33, 34]. EndA is an endonuclease responsible for converting double-stranded DNA (dsDNA) into single-stranded DNA (ssDNA) for cellular uptake and subsequent recombination in the surrounding milieu of pneumococci [35]. The transforming strand is then transported into the cytoplasm in the 3’ to 5’ direction [36–38], whereas the non-transported strand was postulated to be degraded [34]. The fact that EndA is unique among proteins of the uptake apparatus raises the question of whether or not it is already present in cells before they are competent. Furthermore, its subcellular localization in non-competent cells and how it is recruited into the transformation machinery must also be answered. Based on the study by Rosenthal and Lacks [39], the nonspecific EndA sequence represents the first DNA uptake apparatus and was also reported as a membrane-localized pneumococcal endonuclease. Bergé et al. [40] detected the nucleolytic activity of EndA in competent pneumococcal cells during genetic transformation and this nucleolytic activity is indicative of the bacterial competent phase. It could thus be present before the pneumococcal cells switch into a competent state.
In addition, Zhu et al. [41] suggested that EndA was expressed at different concentration levels depending on various factors. Different pneumococcal strains also secrete varying levels and activities of EndA in the growth medium. Therefore, these unique characteristics of EndA facilitate streptococci to establish genetic diversity during a competent state and take part in the organism’s ability to adapt to changing environmental conditions to provide a significant advantage during infection [38].
The pathogenesis of S. pneumoniae is an intricate and dynamic process. The pathogen expresses multiple virulence factors, which can combine to induce invasive pneumococcal disease [42]. The flexibility of S. pneumoniae facilitates responses to evolutionary pressures that provide a significant advantage during infection, such as evasion of host immune defenses or developing antibiotic resistance [43]. In addition, EndA in S. pneumoniae plays a dual role in DNA uptake and contributes to the genetic flexibility that characterizes the pathogen [44].
It is also crucial to understand the neutrophils’ general structure and mechanism since the nucleolytic activity of EndA could facilitate the degradation of NETosis (NETs). Neutrophils are recognized as essential effector cells of innate immunity and critical regulators of both natural and adaptive immune responses [45]. Neutrophils have a short half-life of less than 24 hours [46] and will differentiate and exit the bone marrow before dying in the bloodstream in the absence of infection. When the host becomes infected, tissue-resident macrophages and other sentinel cells emit inflammatory cytokines and chemo-attractants to recruit and stimulate neutrophils. In response to the production of these chemicals, neutrophils exit the bloodstream and penetrate the infected tissues via a selection and integrin-mediated process known as extravasation [46]. Neutrophils are the first effector cells to reach the infection site, thus making them essential in pathogen clearance, recruitment, activation of other immune cells, and tissue repair [46].
Mitochondrial DNA was revealed as a significant structural component of NETs in an early finding reported by Brinkmann et al. [47]. Physically, by using electron microscopy, the NET structure was found to be made up of fibrous DNA stretches with widths ranging from 15 nm to 17 nm and globular protein domains around 25 nm that may aggregate to more extensive threads with diameters exceeding 50 nm [47]. Von Köckritz-Blickwede et al. [48] also confirmed the absence of membrane proteins and cytoplasmic fragments within the NET structures, as well as highlighting the presence of histones as intricate components [48]. Furthermore, these structures are primarily composed of nuclear or mitochondrial DNA, serving as the foundation for antimicrobial peptides, enzymes, and histones, whereas some researchers argue that the contribution of DNA from mitochondria to NETs is minimal [49]. These large extracellular structures provide a physical barrier by increasing the local concentration of antimicrobial effectors and could prevent microbial dissemination [50, 51]; hence, understanding their structure and physiology could provide us with knowledge to avoid the further spread of pathogens upon infection. Microbiological stimuli from bacteria, fungi, viruses, and protozoa are examples of NETosis inducers. Besides, additional chemical factors and reactive oxygen species (ROS) may contribute to NETosis, as listed in Table 1.
Structure-factor stimuli of pathogen-inducing NETs. Depending on the stimulus, neutrophils can become either suicidal (terminal) or vital “zombie” neutrophils
| Group | Species |
|---|---|
| Bacteria | Shigella flexneri [47], Escherichia coli [51], Streptococcus pyogenes [52], Streptococcus pneumonia [53], and Staphylococcus aureus [49, 54, 55]. |
| Viruses | HIV-1 [56], influenza [57], feline leukemia virus [58], and rabbit poxvirus (MYXV) [59]. |
| Fungi | Candida albicans [60], Aspergillus fumigatus [61], and Aspergillus nidulans [62]. |
| Parasite | Leishmania spp. [63], Plasmodium falciparum [64]. |
| Chemicals | Phorbol-12-myristate-13-acetate (PMA) [47], hydrogen peroxide (H2O2) [54], antibodies [61], antigen-antibody complexes [65], microbial components [66], lipopolysaccharide (LPS) [67], M1 protein from Streptococcus pyogenes [68], lipophosphoglycans from Leishmania amazonensis [63], toll-like receptor 4 (TLR4) activated platelets [69], and macrophage-1 (MAC1) integrin receptors [69]. |
Neutrophils are the first immune cells that reach the site of infection and support the clearance of harmful bacteria through numerous processes, including developing NETs, as illustrated in Figure 2. Initially, phagocytosis and degranulation were considered the main mechanisms carried out by neutrophils to eliminate pathogens. As demonstrated in Figure 2, the first sign of NET formation via cell death or suicidal NETosis mechanism is changing the morphology of the nuclease, which loses its characteristic lobulated form. Neutrophil elastase (NE), generally stored in neutrophil granules, translocates into the nucleus during NETosis and cleaves histones by promoting chromatin decondensation [70]. Nuclear membranes and chromatin decondensed into the cytoplasm while the plasma membrane remains intact. Ultimately, the plasma membrane ruptures, leading to the release of NETs. The receptors responsible for initiating NET release and the molecular mechanisms through which ROS drives this process are poorly understood.
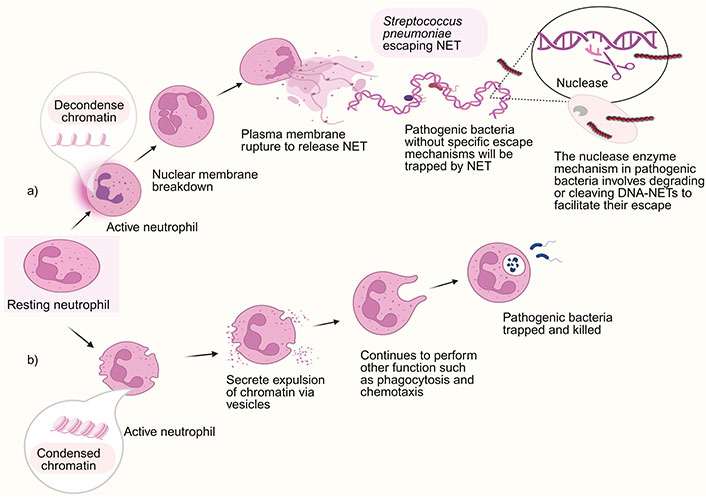
NET formation. Neutrophils employ two primary strategies to combat microbes: phagocytosis and releasing NETs depending on their stimuli. (a) Suicidal/classical NETosis with escaping mechanism of invasive S. pneumoniae that expressed virulent factor; (b) vital NETosis. NET: neutrophil extracellular trap. Created in BioRender. Yusoff, M. (2023) BioRender.com/n72w387
However, certain pathogenic microorganisms possess the capacity to express virulence factors that enable them to successfully establish an infection and evade the host immune system. For example, S. pneumoniae induces NET formation but has evolved virulence mechanisms to avoid NETs. The invasive pneumococcus type TIGR4 expresses EndA, which enables escape from NETs by degrading the extracellular traps, thereby increasing their virulence in vivo [71] (Figure 2a). The evasion of NETs facilitates the dissemination of bacteria from the upper respiratory tract to the pulmonary region and from the lungs to the bloodstream [52].
In contrast, the first mechanism discovered is vital NET formation [72] (Figure 2b) which occurs via slow lytic cell death. A few NETs are produced when neutrophils quickly release their nuclear content by vesicular secretion. Their nuclear content expulsion via vesicles to intact cytoplasts that continue crawling and digesting microbes using phagocytosis or chemotaxis [73]. The significance of these structures in the immune response is highlighted by the description of pathogen compounds that can impede, destroy, and elude the antimicrobial action of NETs (Figure 2) [74].
NETs have been shown to play a physiological protective role in infection sites and inhibit the spread of pathogens. In response to their phagocytic function, NETs provide several benefits to reduce disease spreading by concentrating host antimicrobial agents at infection sites. For example, it has been shown that NETs in staphylococcal skin infections inhibit the penetration of pathogens into the bloodstream [75]. Investigations on S. pneumoniae-infected humans and animals have shown increased NETs within alveolar gaps [76]. Moreover, an in vitro investigation has shown that NETs have vigorous antibacterial activity against S. pneumoniae [53]. These results demonstrated the importance of NETs, particularly in pneumococcal pneumonia.
Despite the variable efficiency of the antimicrobial properties of NETs, some microbes have evolved mechanisms and strategies to reduce the effectiveness of NETs. The pathogenic systems have three functions: deactivate the parts of NETs that function to trap and eliminate pathogens, inhibit the growth of NETs, and create defense mechanisms against the antimicrobial components of NETs [77]. Previous studies have demonstrated the various mechanisms utilized by pathogens to escape from NETs by utilizing the bacterial polysaccharide capsule, formation of biofilms, altering the electric charge of the cell surface, inhibiting ROS formation or inhibition of NET production by peptidase and DNase generation [78].
As part of their defensive strategies, bacteria produce nucleases to protect themselves by breaking down the NETs. This is because the DNA backbone of NETs contains bactericidal molecules such as histones or cathelicidin that can kill entrapped bacteria. Membrane-bound bacterial REases, which are released into the surrounding environment in response to NETs might thus be considered as a protective virulence factor of the bacteria [79]. Numerous S. pneumoniae strains have been shown to produce an endonuclease that renders them entirely immune to NET-induced death and has been shown to compromise the functional integrity of NETs [72]. In his early study, Beiter et al. [73] reported that EndA represents a major nuclease that allows pneumococci to degrade the DNA scaffold of NETs and then escape from NETs efficiently. By using the mouse intranasal model, Beiter et al. [73] showed that during pneumonia, pneumococci travelled more easily from the upper airways to the lungs and into the bloodstream when they managed to escape out of the NETs, contributing to bacterial resistance to innate immunity. Therefore, understanding the endonuclease mechanism in escaping NETs could give a clear perspective on manipulating nuclease secretion, making S. pneumoniae less invasive to humans.
Similarly, Peterson et al. [42] reported that the surface EndA of S. pneumoniae performs two crucial roles in pneumococcal illness. Initially, EndA was responsible for the transport and destruction of DNA during transformation, which adds to the genomic variety of pneumococci. Secondly, as illustrated in Figure 3a, it allows the extracellular breakdown of the DNA component of NETs, which promotes pneumococcal spread and raises the risk of invasive infection [42]. EndA deletion in the study reduced transformation efficiency, potentially impeding the genetic diversity responsible for pneumococcal pathogenicity. Furthermore, in experiments on mice, pneumococci lacking EndA exhibit decreased invasive infection and cannot escape from NETs [72]. Specifically, the previous findings on mutant knock-out for EndA could mitigate the invasion of S. pneumoniae due to the inability to escape through NETs, leading to unsuccessful colonization of S. pneumoniae in the host. Therefore, these would give promising attenuating pneumococcal pathogenesis to increase immunogenicity protection and offer a novel target for controlling pneumococcal disease (see Figure 3b).
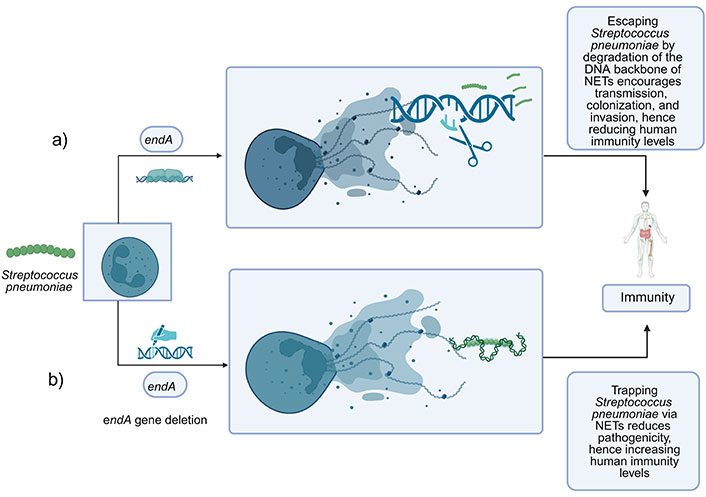
A relationship between EndA in S. pneumoniae towards immunogenicity protection that contributes to immunity level. (a) Wild type S. pneumoniae secreted EndA extracellularly during growth, which can degrade the DNA of NETs hence helping the pathogen establish infection by transmission, colonization, and invasion in the lung; (b) mutant type of S. pneumoniae with deletion of the endA gene are found to have reduced DNA degradation activity in NETs. The endA gene deletion mutant significantly reduced escaping through NETs and in the lung and could increase the immune response against the invading S. pneumoniae. NETs: neutrophil extracellular traps. Created in BioRender. Yusoff, M. (2023) BioRender.com/u26n036
However, excessive NETs have been reported to be associated with reduced clinical stability hazards and increased pneumonia mortality [80]. Higher concentrations of NETs released by activated neutrophils promoted tissue damage, including lung injury [81] and sepsis [82]. The component mechanisms that are responsible for NET-induced tissue damage are NE components and other proteases that induce cell death in multiple cell types [83–87]. Instead of pneumonia, NETosis contributes to developing complications for the infected lung, including bronchial asthma, acute respiratory distress syndrome (ARDS), and chronic obstructive pulmonary disease (COPD). Before the occurrence of the COVID-19 pandemic, a clinical experiment found a link between the severity of community-acquired pneumonia and the content of NETs [80]. Two decades after the discovery of NETs, a significant correlation emerges between the release of NETs and the immune system and various diseases [88].
Since NET formation and elimination were unregulated and led to severe effects, the focus of NET research has changed from innate immune defense to NET involvement in severe consequences [88]. Recent in vitro and in vivo experiments using animal models demonstrated the crucial role of NETs in pathogenesis and increasing morbidity and mortality [88]. Multiple organ failure and poor prognoses have been linked to high levels of circulating NETs in sepsis patients [89–91]. In addition, NETs have been linked to pathologic changes in autoimmune and autoinflammatory illnesses [92, 93] due to the circulating free DNA (cfDNA) exacerbating inflammation by inducing TNF-α mRNA [94]. At the same time, it has been demonstrated that these inflammatory substances increase NETosis and pathology in these patients without causing infection [95]. For example, the deposition of monosodium urate crystals in the joints is an autoinflammatory disease called gout that stimulates an immune response by attracting leukocytes to induce NETs, hence promoting inflammation [95]. Undoubtedly, excessive NET formation and poor elimination mechanisms of NETs can cause extreme tissue damage and pathology that lead to inflammatory and autoimmune pathologies [96].
There are still many unsolved essential concerns about the exact functional roles that NETs play in the etiology of various illnesses. The release of enzymes and other proteins during NETosis causes tissue injury, and EndA produced by many strains of S. pneumoniae, enables them to escape NETs by degrading their functional integrity. Buchanan et al. [52] found that increasing bacterial migration from the upper airways to the lungs was linked to nuclease digestion of the DNA scaffold of NETs, possibly contributing to a 20–30% increase in cases of invasion into the bloodstream. However, reducing this virulence factor of pathogens does not address all issues, as it is crucial to consider all possible mechanisms and their consequences. Therefore, understanding the role of bacterial nucleases in degrading NETs is important, as they can balance NET secretion and potentially serve a protective role in managing infections.
Above all, the exploration of nucleases becomes attractive. Researchers have come out with various findings to understand the mechanism of nucleases as one of the NETs’ therapeutics [97, 98]. In addition, EndA is a desirable target for antimicrobial therapies due to its function as a virulence component in pneumococcal infection. A previous study by Peterson et al. [42], using a high throughput screening assay, uncovered six small molecules that were potent EndA inhibitors. These small-molecule inhibitors were shown to be a promising starting point for the development of pharmacologic EndA probes. By doing this, the virulence of pneumococcal bacteria might be addressed, and it could be identified as a target for novel compounds to fight pneumococcal infection [42].
DNase I and anti-histone antibodies have also become exciting targets for researchers instead of drugs aimed at destroying NETs. In the beginning, the recombinant DNase I significantly facilitates the course of ARDS [98] and COPD [99] in animal model studies. Since then, it has been effectively applied to practically all models of NETosis-related disorders. For instance, Pulmozyme (Dornase alfa), a recently discovered highly pure solution of recombinant human deoxyribonuclease I (rhDNase), was commercialized by Genentech [100]. This enzyme’s selectively cleaving DNA mechanism becomes a key factor of success due to hydrolyzing the DNA in the sputum/mucus of cystic fibrosis patients, reducing lung viscosity and promoting clearance of secretions [101].
Meanwhile, the impact of rhDNase on NETs has been assessed in various investigations. According to some studies, a decrease in NETosis will decrease neutrophil infiltration and the inflammatory response [102, 103]. Findings from an alternative approach show that in septic circumstances, early concurrent treatment with DNase I and antibiotics improved survival and decreased bacteremia and organ dysfunction [104]. Kaplan et al. [105] suggested that possible combination therapy will assist in controlling NETosis. These enzymes may significantly impact human medicine in the future by providing novel therapeutic options for conditions where multi-drug resistance had otherwise caused treatment failure. However, further studies are needed to validate whether these effects are due primarily to NET inhibition.
EndA is a significant virulence factor in streptococci, enabling the establishment of genetic diversity and enabling S. pneumoniae to initiate invasive pneumococcal infections. Surface EndA facilitates the degradation of the DNA scaffold of NETs, enhancing the ability of the pathogenic bacteria to escape from these traps. Deleting the endA gene in pneumococci reduces invasive infections and enhances immunogenicity protection. However, an overabundance of NETs is significant in developing infectious and inflammatory disorders. A novel approach to address this issue involves investigating the pathophysiological significance of NETosis and its potential involvement in pneumonia development. NET-targeted interventions, hydration therapy, antibiotics, antivirals, and conventional medications might require integration for therapeutic effectiveness. From this perspective, nucleases such as EndA exhibit various bacterial pathogenesis and virulence functions while presenting a potential therapeutic target. One possible outcome involves the development of drugs or vaccines that can effectively balance the activity of nucleases. Targeting EndA in S. pneumoniae could enhance NET formation stability, strengthen immune defense mechanisms, and improve treatment outcomes.
EndA: endonuclease A
HGT: horizontal gene transfer
MTase: methyltransferase
NETs: neutrophil extracellular traps
REase: restriction endonuclease
R-M: restriction-modification
ROS: reactive oxygen species
MY: Conceptualization, Writing—original draft, Writing—review & editing. CCY and MA: Validation, Supervision, Writing—review & editing. MHN: Writing—review & editing. All authors read and approved the submitted version.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
This work was funded by the Universiti Sultan Zainal Abidin Research Centre Grant [UniSZA/2020/LABMAT/03] to support research projects in the Centre for Research in Infectious Diseases and Biotechnology (CeRIDB) at the Faculty of Medicine. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
© The Author(s) 2024.
Copyright: © The Author(s) 2024. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
View: 3487
Download: 50
Times Cited: 0
