Affiliation:
1Department of Internal Medicine, Eastern Virginia Medical School, Norfolk, VA, USA
ORCID: https://orcid.org/0009-0003-7994-1371
Affiliation:
2Department of Internal Medicine, Atrium Health, Charlotte, NC, USA
ORCID: https://orcid.org/0000-0001-9806-0615
Affiliation:
3Department of Internal Medicine, Virginia Commonwealth University, Richmond, VA, USA
ORCID: https://orcid.org/0000-0002-8441-0132
Affiliation:
1Department of Internal Medicine, Eastern Virginia Medical School, Norfolk, VA, USA
ORCID: https://orcid.org/0000-0002-0597-0247
Affiliation:
4Department of Internal Medicine, Case Western Reserve University, Cleveland, OH, USA
ORCID: https://orcid.org/0000-0001-9314-5361
Affiliation:
5Division of Gastroenterology and Hepatology, Case Western Reserve University, Cleveland, OH, USA
ORCID: https://orcid.org/0000-0003-3772-2616
Affiliation:
6Division of Gastroenterology and Hepatology, Eastern Virginia Medical School, Norfolk, VA, USA
Email: dajevms@aol.com
ORCID: https://orcid.org/0000-0002-8737-0711
Explor Med. 2025;6:1001275 DOI: https://doi.org/10.37349/emed.2025.1001275
Received: November 01, 2024 Accepted: December 17, 2024 Published: January 19, 2025
Academic Editor: Zeneng Wang, Cleveland Clinic, USA
The article belongs to the special issue Gut Microbiota Derived Metabolites and Chronic Inflammatory Diseases
The gut microbiota, a complex ecosystem of microorganisms, plays an essential role in maintaining immune and metabolic homeostasis. Disruption of this microbial balance, known as dysbiosis, has been increasingly implicated in the pathogenesis of chronic inflammatory conditions, including cardiovascular, gastrointestinal, and autoimmune diseases, as well as metabolic disorders such as diabetes and obesity. A crucial mechanism through which the gut microbiota exerts its effects on host physiology is via the production of bioactive metabolites. These metabolites, including short-chain fatty acids, bile acids, and tryptophan derivatives, are key in modulating immune responses and regulating metabolic functions. Dysbiosis disrupts the production and function of these metabolites, thereby contributing to immune dysregulation, chronic inflammation, and disease progression. This review examines the role of gut microbiota-derived metabolites in chronic inflammatory diseases, with a focus on their immunomodulatory and metabolic effects. A deeper understanding of these mechanisms may open the way for novel therapeutic strategies aimed at restoring immune homeostasis and mitigating the global burden of chronic inflammatory diseases.
Chronic inflammatory diseases encompass a wide range of conditions, including autoimmune disorders, cardiovascular, gastrointestinal (GI), chronic kidney and liver diseases, all of which represent a significant global health burden [1]. Despite advancements in early diagnosis and treatment, which have improved patient outcomes, the prevalence of these conditions continues to rise. According to the 2018 National Health Interview Survey (NHIS), more than half (51.8% or approximately 129 million) of U.S. adults have been diagnosed with at least one major chronic condition, with 27.2% experiencing multiple conditions [2]. This growing prevalence is largely driven by demographic shifts and lifestyle factors.
The pathogenesis of chronic inflammatory diseases involves a complex interaction of genetic and environmental factors that contribute to epigenetic modifications and disruptions in cellular signaling pathways [3]. Among these, the gut microbiome has recently emerged as a critical mediator serving as a dynamic interface between environmental factors and host physiology [4, 5]. Composed predominantly of the bacterial phyla Firmicutes and Bacteroidetes, the gut microbiome plays an essential role in maintaining metabolic balance, modulating immune responses, preserving intestinal barrier integrity, digesting nutrients and metabolites, and influencing cellular signaling pathways [6, 7].
Environmental factors such as diet, geographical location, delivery mode (vaginal versus cesarean section), and lifestyle behaviors significantly shape the gut microbiome and its downstream effects on the immune system [8]. Diet, in particular, has a profound impact on microbial composition and function. High-fiber diets encourage the growth of beneficial microbial species and the production of anti-inflammatory metabolites, while diets high in saturated fats and refined sugars promote a pro-inflammatory microbiome [9, 10]. Geographical location and delivery mode also influence the initial colonization and diversity of the gut microbiota, further affecting immune responses [11, 12]. Additionally, physical activity and exposure to medications, such as antibiotics, can modify the gut microbiome, altering the production of metabolites critical for immune regulation [13, 14].
The influence of these environmental factors can disrupt the balance of the gut microbiota, leading to dysbiosis. Dysbiosis is characterized by an increase in pro-inflammatory species and a decrease in anti-inflammatory ones, triggering widespread physiological disturbances, systemic inflammation, and the development of various diseases [4, 6, 15]. Dysbiosis not only alters the composition of the gut microbiota but also disrupts the production of key metabolites, exacerbating inflammatory processes and impairing immune regulation.
Understanding how microbial imbalances contribute to immune dysfunction and chronic inflammation is essential for developing targeted therapeutic strategies. This review will explore the classification of microbial metabolites, the related mechanisms of immune modulation, and involvement in the pathogenesis of chronic inflammatory diseases.
The gut microbiota plays a fundamental role in regulating innate and adaptive immune responses, helping maintain immune homeostasis. These effects occur through direct interactions between commensal microorganisms and the host’s immune cells, as well as indirect modulation by gut microbiota-derived metabolites and signaling molecules [16]. Among these metabolites, short-chain fatty acids (SCFAs), bile acids (BAs), microbial-derived vitamins, and tryptophan catabolites are particularly significant [17] (Figure 1). These bioactive compounds exert local and systemic immunomodulatory effects by engaging specific receptors, including G-protein-coupled receptors (GPRs), nuclear receptors, and pattern recognition receptors (PRRs). Such interactions influence immune cell functions, cytokine production, and immune balance, thereby impacting physiological processes both within and beyond the GI tract [18].
Gut microbiota-derived metabolites can be broadly categorized into three major groups based on their origin and function, all of which play crucial roles in immune regulation and are implicated in various diseases when their production is altered (Table 1).
Gut microbiota-derived metabolites and their immunomodulatory roles
| Metabolite type | Immunomodulating property | Associated diseases | References |
|---|---|---|---|
| Metabolic end products | |||
| Short-chain fatty acids (SCFAs) | Support gut barrier integrity, reduce pro-inflammatory cytokines, promote Treg differentiation, neutrophil migration, and B cell class switching | Metabolic syndrome, cardiovascular disease, inflammatory bowel disease, colorectal cancer, and chronic kidney disease | [19–24] |
| Tryptophan and indole derivatives | Regulate gut barrier, motility, and hormone secretion; modulate immune responses via AhR receptor, promoting immune tolerance | Inflammatory bowel disease, irritable bowel syndrome, obesity, colorectal cancer | [18, 23, 25, 26] |
| De novo products | |||
| Branched-chain amino acids (BCAAs) | Crucial for protein synthesis, cell growth, and energy metabolism | Type 2 diabetes, obesity, cardiovascular disease, colorectal cancer | [17, 23, 27, 28] |
| Polyamines (PAs) | Maintain intestinal barrier, reduce oxidative stress, inhibit inflammatory cytokine production | Colorectal cancer, other tumors | [23, 29–31] |
| Bacterial-derived vitamins | Support metabolic processes, regulate DNA synthesis, energy metabolism, immune cell function | Cardiovascular disease | [18, 32, 33] |
| Modified host metabolites | |||
| Secondary bile acids (BAs) | Modulate activity of innate immune cells (macrophages, dendritic cells), regulate lipid and glucose metabolism | Metabolic dysfunction-associated steatotic liver disease, inflammatory bowel disease, colorectal cancer | [18, 34–36] |
AhR: aryl hydrocarbon receptor; Tregs: regulatory T cells
These metabolites are produced by intestinal flora through the consumption of luminal dietary components.
SCFAs, such as acetate, butyrate, and propionate, are derived from the fermentation of dietary fibers. They maintain gut barrier integrity by upregulating Claudin-1 expression and acting as an energy source for intestinal epithelial cells [19]. SCFAs also stimulate mucus secretion and promote M2 macrophage polarization, enhancing pathogen defense [20]. They also modulate systemic immune responses by interacting with GPRs on immune cells [21–23]. Alterations in SCFA levels, as seen in dysbiosis, have been implicated in the development of inflammatory bowel disease (IBD), metabolic syndrome, cardiovascular diseases (CVDs), chronic kidney disease (CKD), and colorectal cancer [24]. Reduced SCFA production disrupts gut homeostasis, promoting chronic inflammation and metabolic dysregulation.
Tryptophan and indole derivatives, produced through the bacterial metabolism of tryptophan, play an important role in regulating gut barrier function by enhancing epithelial integrity and maintaining tight junctions. They also influence gut motility and hormone secretion, including the stimulation of glucagon-like peptide-1 (GLP-1), which affects insulin secretion, appetite suppression, and gastric emptying [18, 23]. Additionally, indole derivatives modulate immune responses through interactions with the aryl hydrocarbon receptor (AhR), a critical regulator of mucosal immunity. This interaction impacts local and systemic immune functions, contributing to immune tolerance and protection against inflammation [25, 26]. Disruptions in indole derivative production have been linked to ulcerative colitis (UC), Crohn’s disease (CD), irritable bowel syndrome (IBS), liver fibrosis, and neurodegenerative diseases such as Parkinson’s disease, as well as colorectal cancer [26].
Branched-chain amino acids (BCAAs), including leucine, isoleucine, and valine, are vital for protein synthesis, energy metabolism, and cellular growth. BCAAs also play a role in modulating immune cell functions, especially during bacterial invasion, where there is increased demand for them to sustain the high metabolic activity of immune cells like T-cells [17, 23]. Dysregulation of BCAA synthesis and degradation has been associated with conditions such as type 2 diabetes, obesity, CVDs, and colorectal cancer [23, 27, 28].
Polyamines (PAs), including putrescine, spermidine, and spermine, are small polycationic molecules essential for gene regulation, cell proliferation, differentiation, and stress responses [29]. These molecules are key in maintaining intestinal barrier function, reducing oxidative stress, and inhibiting inflammatory cytokine production [23]. Dysregulated polyamine metabolism may contribute to colorectal cancer and other tumors by supporting tumor growth [30, 31].
Bacterial-derived vitamins, such as vitamins K and B (e.g., B12, B2, and B9), are essential for various metabolic processes such as DNA synthesis, energy metabolism, and red blood cell production [32]. Additionally, these vitamins support immune cell function and maintain intestinal barrier integrity, contributing to immune homeostasis. For example, vitamin B2 (riboflavin) influences the activity of mucosal-associated invariant T (MAIT) cells, which play a role in the body’s response to microbial antigens. In contrast, vitamin B9 (folate) and B12 (cobalamin) are crucial for regulating immune responses by promoting T-cell proliferation and ensuring the proper functioning of the methylation cycle, essential for DNA synthesis and the activation of immune cells [18, 23].
Certain metabolites produced by the host, such as primary BA, are modified by gut bacteria into secondary BA [18, 34].
Secondary BAs interact with specific immune receptors like the farnesoid X receptor (FXR) and Takeda GPR 5 (TGR5), modulating the activity of innate immune cells. Additionally, they play a significant role in regulating lipid and glucose metabolism [35, 36]. Altered BA metabolism is associated with metabolic dysfunction-associated steatotic liver disease (MASLD), IBD, and colorectal cancer [18].
Figure 1 outlines the importance of the gut microbiota in regulating immune homeostasis. Indeed, the gut microbiota shapes both innate and adaptive immune responses to maintain homeostasis through direct interactions with the host immune cells and via indirect ways, which consider the production of metabolites by the microbiota. Key metabolites-SCFAs, BAs, microbial-derived vitamins, and tryptophan catabolites-exert local and systemic immunomodulatory effects.
The immunomodulatory effects of gut microbiota-derived metabolites are mediated through specific receptor interactions that influence cellular signaling pathways, immune cell function, and inflammatory responses.
GPRs are transmembrane proteins that detect extracellular metabolites, such as SCFAs and BAs, activating intracellular signaling cascades through G proteins and β-arrestins. These receptors are key players in metabolic regulation and immune modulation.
GPR41, GPR43, and GPR109A respond to SCFAs, promoting T-cell differentiation, cytokine production, B cell activation, and modulating macrophage and dendritic cell functions to support immune balance and gut motility [37, 38]. GPR35 interacts with tryptophan-derived metabolites, like kynurenic acid (KYNA), supporting gut barrier integrity and exerting anti-inflammatory effects, particularly in invariant natural killer (iNK) cells [39, 40]. Conversely, 5-hydroxyindoleacetic acid (5-HIAA), released by platelets and mast cells, activates GPR35 to recruit dendritic cells, neutrophils, and eosinophils, promoting immunoglobulin A (IgA) production by plasma cells, but also contributing to inflammation [41]. GPR131, also known as TGR5, is activated by BAs and plays a role in inflammation by suppressing pro-inflammatory cytokines and promoting an anti-inflammatory phenotype in macrophages [42].
AhR is a nuclear receptor activated by tryptophan-derived metabolites. AhR modulates immune responses by influencing the differentiation of T cells [Th17 and regulatory T cells (Tregs) cells] and the function of innate lymphoid cells, macrophages, and dendritic cells. It also promotes epithelial renewal and gut barrier integrity, playing a central role in maintaining intestinal homeostasis and regulating inflammation [37, 43].
The pregnane X receptor (PXR), vitamin D receptor (VDR), and FXR are nuclear receptors that play significant roles in immune regulation. Activated by BAs and other ligands, these receptors modulate immune responses by reducing pro-inflammatory signaling, influencing dendritic cell activity, and regulating the balance of T-cell subsets. VDR and FXR promote the differentiation of Tregs while inhibiting the differentiation of pro-inflammatory Th17 cells, contributing to immune tolerance and homeostasis [37, 44].
PPRs, including Toll-like receptors (TLRs) and nucleotide-binding oligomerization domain-like receptors (NLRs), play a crucial role in linking gut microbiota-derived metabolites to immune regulation. TLRs detect microbial-associated molecular patterns (MAMPs), including lipopolysaccharides (LPS) and peptidoglycans, to initiate signaling pathways that result in the production of inflammatory and anti-inflammatory cytokines [16]. NLRs, including NLRP6, recognize bacterial fragments and regulate inflammasome activity, essential for balancing cytokine secretion and maintaining gut microbiota composition [45].
IBD, which includes CD and UC, exemplifies a chronic inflammatory condition with complex etiology involving genetic, environmental, and microbial factors. Among the various influences, metabolites derived from gut microbiota, such as SCFAs, hydrogen sulfide (H2S), and BAs, are crucial in the disease’s pathogenesis and management, primarily through their roles in modulating immune responses and maintaining intestinal integrity [46].
SCFAs are essential for maintaining the epithelial barrier’s integrity, which is vital for overall intestinal health. In conditions of dysbiosis, a significant decrease in SCFA levels contributes to barrier weakening, thereby facilitating increased bacterial translocation that perpetuates the inflammatory cycles central to IBD development [47]. Furthermore, SCFAs enhance the development of B and Tregs, mitigating the pro-inflammatory states frequently observed in dysbiosis [48, 49]. However, the microbiomes of IBD patients often show a diminished presence of SCFA-producing bacteria, leading to elevated levels of pro-inflammatory cytokines as macrophages adopt a more inflammatory phenotype [49, 50].
Similarly, H2S, a gaseous metabolite produced from cysteine and sulfate by numerous bacterial species including cystein catabolic bacteria (e.g., Fusobacterium) and sulfate-reducing bacteria (e.g., Desulfovibrio), exhibits a dual role in gut health. Under physiological conditions, H2S contributes to intestinal homeostasis by enhancing the mucus barrier, stabilizing microbial biofilms, and regulating immune responses to limit pro-inflammatory cytokine activity such as tumor necrosis factor-α (TNF-α), and interferon-γ (IFN-γ) [51]. However, recent evidence suggests that both excessive and insufficient levels of H2S can compromise epithelial integrity, promote oxidative stress, and aggravate inflammation [52, 53]. In IBD, dysbiosis often leads to an overgrowth of sulfate-reducing bacteria and reduced activity of H2S-detoxifying enzymes, exacerbating inflammation and epithelial damage [54]. Although H2S’s precise role in IBD pathogenesis remains an active area of research, current evidence highlights its significance in disease progression.
The role of the microbiome extends to the modulation of immune responses through BAs as well. Produced by the liver, BAs are modified by the gut microbiome, impacting immune regulation by affecting T cell activity to either promote or reduce inflammation [49]. Significantly, IBD patients often exhibit reduced levels of these modified BAs, suggesting an important mechanism by which dysbiosis may influence the regulation of the intestinal immune system [55].
Given the association between dysbiosis and intestinal inflammation, strategies to modulate the microbiome represent promising therapeutic avenues for altering the course of IBD. Probiotics, which confer health benefits, have been associated with improved outcomes in IBD, helping reduce disease recurrence and promote remission in both CD and UC patients [56]. Additionally, fecal microbiota transplantation (FMT) has gained traction as a treatment, for recurrent Clostridium difficile infection and IBD, which has been linked to improved remission rates. The success of FMT, however, may depend on various factors, including the donor’s microbial composition, the frequency of transplantation, and the timing relative to the disease course [56].
IBS is a functional GI disorder characterized by symptoms such as abdominal pain, bloating, and altered bowel habits that significantly impact patients’ lives. Although the precise etiology of IBS is not well understood, recent evidence has emphasized the substantial role of the gut microbiome in its pathogenesis [57]. The brain-gut axis (BGA), a bidirectional pathway connecting the nervous system with the GI tract, plays a crucial role in this context [58]. It encompasses pathways involved in immune system regulation, hormonal balance, and the autonomic nervous system [59]. Gut dysbiosis can disrupt the BGA, leading to altered intestinal motility and visceral hypersensitivity, common in IBS [60].
Patients with IBS often show a microbial profile characterized by a decrease in beneficial bacteria such as Bifidobacterium and Faecalibacterium prausnitzii, and an increase in potentially harmful bacteria like Enterobacteriaceae and Ruminococcus gnavus [61, 62]. This shift is associated with a decreased production of microbial metabolites, including SCFAs, which are crucial for maintaining intestinal barrier function and modulating immune responses [63]. The reduction in SCFAs may impair these protective mechanisms, contributing to the pathophysiology of IBS [47] (Figure 2).
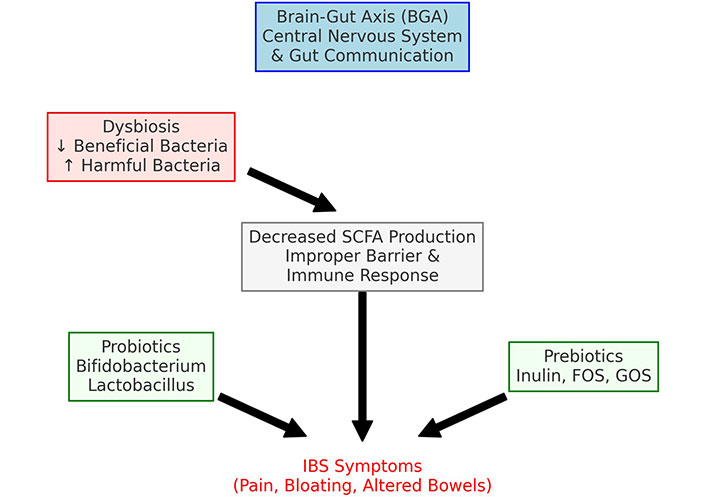
Therapeutic pathways for irritable bowel syndrome. SCFA: short-chain fatty acid; FOS: fructooligosaccharides; GOS: galactooligosaccharides
The link between the microbiome and IBS has advanced our understanding of the disorder and opened new therapeutic avenues. Probiotics have been shown to alleviate IBS symptoms effectively. Studies indicate that specific strains such as Bifidobacterium spp. and Lactobacillus spp. can significantly improve GI as well as psychological symptoms, suggesting that these probiotics positively impact the BGA [64]. Additionally, prebiotics—non-digestible fibers that promote the growth of beneficial microbes—have proven effective. Compounds like inulin, fructooligosaccharides (FOS), and galactooligosaccharides (GOS) have been shown to enhance levels of beneficial bacteria and reduce IBS symptoms [65, 66].
While considerable progress has been made in leveraging the microbiome for managing IBS, further research is necessary. More detailed characterization of the IBS-associated microbiome could lead to targeted probiotic therapies that specifically address disruptions in the BGA, potentially improving both GI and psychological symptoms. Continued efforts to elucidate the complex interactions between the microbiome and IBS are crucial for developing more effective and specific therapeutic interventions for patients with this disorder.
Obesity/Metabolic syndrome reflects systemic disorders characterized by impaired metabolic regulation, closely linked to insulin resistance, chronic inflammation, and dysregulated lipid metabolism. The role of gut microbiota-derived metabolites, particularly BCAAs, SCFAs, and tryptophan metabolites, has emerged as a critical factor in these conditions. Recent research highlights these metabolites’ profound influence on metabolic pathways, revealing their central role in modulating insulin sensitivity, inflammatory responses, and overall energy balance.
BCAAs have been found to regulate lipid metabolism and insulin sensitivity [67]. There is a dynamic, circular relationship between BCAAs and the gut microbiota. Studies in mice have shown that supplemented diet rich in BCAAs promotes a healthier microbial balance, with increases in beneficial bacteria like Bifidobacterium and Akkermansia and reductions in Enterobacteriaceae [68]. However, elevated BCAAs levels are also associated with insulin resistance and obesity [69]. In genetically obese mice, excessive BCAAs accumulation can directly induce insulin resistance [70]. Moreover, the gut microbiota further influences BCAA metabolism by producing and utilizing these amino acids. For instance, Bacteroides vulgatus and Prevotella copri are associated with BCAAs production, which correlates with obesity and insulin resistance. Conversely, lower amounts of bacteria able to use BCAAs, such as Butyrivibrio crossotus and Eubacterium siraeum, are seen in individuals with insulin resistance as BCAA levels are elevated [71].
Tryptophan, an amino acid found in foods such as oats, milk, cheese, poultry, and fish, plays a crucial role in protein synthesis and serves as a precursor for essential metabolites. Tryptophan follows three primary metabolic pathways: serotonin, kynurenine, and a third pathway involving gut microbiota, which metabolizes tryptophan into indoles and their derivatives [72, 73]. These indole metabolites act as ligands for the AhR, triggering the transcription of key pathways that regulate lipid metabolism and glucose homeostasis [74]. Gut microbial dysbiosis, characterized by impaired AhR ligand production, is a critical factor in the development of metabolic syndrome [75]. Impaired activation of the AhR pathway leads to decreased production of GLP-1 and interleukin-22 (IL-22), increasing intestinal permeability and enhancing gram-negative bacteria LPS translocation. This mechanism contributes to the systemic elevated inflammation commonly observed in individuals with obesity and metabolic syndrome [76].
Diabetes mellitus is primarily driven by a combination of genetic, environmental, and lifestyle factors that disrupt glucose metabolism. In type 2 diabetes mellitus (T2DM), this disruption is characterized by insulin resistance, where the body’s cells become less responsive to insulin, coupled with inadequate insulin production by the pancreas. Gut microbial metabolites play a role in the progression and potential mitigation of T2DM through various mechanisms, including the regulation of insulin sensitivity, inflammation, and glucose homeostasis.
Research into gut microbial metabolism has revealed that resistant starch (RS) metabolites improve glucose homeostasis by regulating the secretion of gut hormones. The production of SCFAs through microbial metabolism of RS can stimulate the release of GLP-1 and peptide YY (PYY) by binding to GPRs on intestinal enteroendocrine cells (EECs) [77]. This activation of ECCs promotes their differentiation and triggers the release of GLP-1 and PYY through Gα signaling pathways [78]. GLP-1 stimulates insulin secretion, inhibits glucagon release, slows gastric emptying, and reduces appetite—all of which improve glucose regulation and thwart the development or progression of insulin resistance [79]. Similarly, PYY contributes to appetite regulation, slows gastric emptying, and supports glucose homeostasis [80].
Beyond hormone regulation, butyrate modulates glucose homeostasis via epigenetic mechanisms, notably by inhibiting of histone deacetylase (HDAC) [81]. By inhibiting HDAC, butyrate influences gene expression linked to glucose metabolism [82]. Studies in mice have shown that HDAC activation reduces glucose uptake in skeletal muscle and liver cells by suppressing the expression of glucose transporter type 4 (GLUT4) and promoting gluconeogenesis in the liver via the hepatocyte nuclear factor 4 alpha (HNF4A) pathway [83]. A reduction in butyrate levels has been associated with increased HDAC activity in the colon, impairing glucose regulation [84]. Supplementation with butyrate via RS-rich foods to augment epigenetic regulation and improve glucose homeostasis could be a novel treatment strategy for T2DM.
Butyrate has also been shown to enhance pancreatic β-cell function. In vitro studies demonstrate that butyrate supports β-cell metabolism and viability by preventing inflammatory cytokine-induced dysfunction. This occurs through the downregulation of specific gene expressions linked to inflammation [85]. Further research has shown that butyrate suppresses nuclear factor kappa-B (NF-κB), thereby preventing IL-1β-induced inflammatory gene expression and preserving pancreatic β-cell function, crucial for maintaining proper glucose regulation [86].
However, not all gut microbial metabolites are beneficial. Some, such as LPS and trimethylamine-N-oxide (TMAO), have been implicated in promoting insulin resistance and T2DM. Increased intestinal permeability in T2DM patients allows LPS to enter the bloodstream, triggering TLR4, an inflammatory marker that disrupts insulin signaling and glucose homeostasis [87]. Similarly, elevated TMAO levels are associated with insulin resistance by activating inflammatory pathways that impair insulin signaling [88]. Additionally, BCAAs can interfere with the insulin receptor signaling cascade, further promoting insulin resistance [89].
MASLD is characterized by hepatic steatosis and is closely linked to metabolic disorders such as obesity and insulin resistance. As in other metabolic disorders, gut microbiota metabolites, namely those of vitamins, proteins, and BAs, act as critical regulators in the pathogenesis of this form of steatotic liver disease (Table 2).
Gut microbiota-derived metabolite regulation of MASLD
| Metabolites | Gut microbes | Functions |
|---|---|---|
| Branched-chain amino acids (valine, leucine, isoleucine) | Bifidobacterium, Clostridium sporogenes, Prevotella copri, Phocaeicola vulgatus |
|
| Short-chain fatty acids (acetate, propionate, butyrate) | Bifidobacterium, Bacteroidetes, Campylobacter jejuni, Coprococcus, Clostridium, Roseburia, Staphylococcus aureus, Faecalibacterium |
|
| Choline metabolites (choline, trimethylamine, trimethylamine N-oxide) | Bifidobacterium, Firmicutes, Actinobacteria, Faecalibacterium prausnitzii, Proteobacteria |
|
| Bile acid metabolites (cholic acid, deoxycholic acid, ursodeoxycholic acid) | Bifidobacterium, Lactobacillus, Enterobacter, Bacteroides, Clostridium |
|
GPR: G-protein-coupled receptors; MASLD: metabolic dysfunction-associated steatotic liver disease
Choline, an essential vitamin, undergoes microbial metabolism in the gut, producing trimethylamine (TMA), which is then oxidized in the liver to form TMAO. The gut microbiota composition significantly influences TMA production, and dysbiosis can lead to fluctuations in TMA levels [90]. Research has demonstrated that TMAO contributes to MASLD progression by increasing oxidative stress [91]. It also promotes the expression of proteins associated with the unfolded protein response, leading to dysregulated lipid metabolism in hepatocytes and further advancing the disease [92]. Lastly, TMAO suppresses key enzymes involved in BA synthesis—CYP27A1 and CYP7A1—thereby reducing BA production. BAs serve as signaling molecules that modulate lipid and glucose metabolism, and their dysregulation can lead to increased hepatic triglyceride levels, promoting the development of hepatic steatosis [93].
BCAAs are metabolized in the liver into branched-chain ketoacids (BCKAs), which enter the tricarboxylic acid (TCA) cycle and serve as signaling molecules in hepatocytes. Elevated BCAA levels have been linked to MASLD, likely due to their role in enhancing mitochondrial lipid β-oxidation [94]. Gut dysbiosis, often caused by high-calorie, high-fat diets, has been associated with increased absorption and circulation of BCAAs, exacerbating MASLD [95]. Contrary to BCAA, the metabolism of certain aromatic amino acid derivatives serves a beneficial role in hepatic function. For instance, tryptophan is metabolized by the gut microbiota into indole-3-acetic acid (IAA), which has demonstrated anti-inflammatory effects in the liver, potentially halting the progression of hepatic steatosis in animal models [96]. Indole supplementation in animals has been shown to reduce hepatic steatosis and inflammation by inhibiting the NF-κB signaling pathway. Importantly, Bifidobacterium bifidum plays a significant role in producing IAA from tryptophan, suggesting that promoting beneficial gut bacteria may help in MASLD treatment [97].
In addition to the metabolism of lipids, BA and their metabolites are intricately linked to liver function. Patients with MASLD show increased levels of BA-metabolizing bacteria such as Lactobacillus, Bifidobacteria, Enterobacter, and Clostridium, which produce a variety of BA derivatives. One metabolite, glycine, has been shown to improve steatosis and insulin resistance when supplemented in rats with diet-induced hepatic steatosis [98]. Similarly, taurine, which conjugates with BA, has demonstrated protective effects against MASLD by improving histological steatosis and reducing oxidative stress and fatty acid synthesis [99]. Disruptions in BA metabolism, therefore, play an important role in MASLD pathogenesis, and restoring balance through targeted interventions could be a potential therapeutic strategy [100].
CVD, including coronary artery disease (CAD) and dyslipidemia, arises from a multifactorial interaction of genetic, lifestyle, and environmental factors. Recently, the role of intestinal microbiota and its metabolites in the development and progression of CVD has garnered significant attention, though this remains an emerging area of research requiring further validation [101].
BAs are synthesized from cholesterol in the liver and secreted into the intestine to promote fat emulsification and lipid absorption. Beyond their role in digestion, BAs also act as signaling molecules by binding to receptors such as TGR5 and FXR [102]. FXR, expressed in the intestines, regulates cholesterol homeostasis by promoting the expression of small heterodimer partner (SHP) and fibroblast growth factor (FGF) 15, which modulate cholesterol excretion. Activation of FXR leads to enhanced reverse cholesterol transport, inhibition of intestinal cholesterol absorption, and subsequent reductions in triglyceride and cholesterol levels [103]. Intercept 767 (INT-767), a dual FXR and TGR5 BA receptor agonist, have shown promise in reducing atherosclerosis in murine models, lower LDL-cholesterol, and suppress the expression of aortic inflammatory cytokines by inhibiting the NF-κB pathway [104]. Given the gut microbiome’s influence on BA metabolism, changes in microbial composition could directly impact the development and progression of CAD [105].
TMAO, a gut microbial metabolite derived from choline-rich foods, has been associated with atherosclerosis [106, 107]. Elevated circulating TMAO levels promote foam cell aggregation and atheromatous plaque formation, as seen in mice fed choline-rich diets [108, 109]. Further studies by Ma et al. [110] revealed that TMAO promotes monocyte adhesion by upregulating vascular cell adhesion molecule-1 (VCAM-1) through the activation of protein kinase C (PKC) and NF-κB, leading to endothelial dysfunction and accelerating atherosclerosis progression. Moreover, elevated levels of TMAO interfere with myocardial mitochondrial repair and exacerbate the severity of myocardial infarction [111]. Contribute to atherosclerotic thrombosis by regulating intracellular calcium release via adenosine diphosphate (ADP), collagen, and thrombin, thereby increasing platelet responsiveness [111]. While these findings suggest a potential role for TMAO in CVD, the evidence is largely preclinical, and robust interventional studies are needed to confirm its clinical relevance [112].
SCFAs, produced by gut microbial fermentation of dietary fibers and starches, are another metabolites that influence cardiovascular health. The primary SCFAs contribute up to 10% of the body’s energy and serve as signaling molecules that modulate blood pressure, cardiovascular function, inflammation, and metabolism [113, 114]. SCFAs exert anti-inflammatory effects by binding to GPRs on epithelial, immune, and adipose cells. Notably, propionate and butyrate inhibit NF-κB and TNF signaling pathways, reducing the expression of adhesion molecules like VCAM-1 and intercellular adhesion molecule-1 (ICAM-1), thereby mitigating atherosclerosis risk [115, 116]. The therapeutic potential of SCFA supplementation to slow CAD progression is a promising area for further research.
CKD is a progressive condition marked by the gradual decline of kidney function, often leading to end-stage renal disease (ESRD). Its etiology involves genetic predispositions, environmental influences, and lifestyle factors. Recently, the gut microbiota has emerged as a significant contributor to CKD. The metabolites produced by the gut microbiota can directly or indirectly impact kidney function, making the gut-kidney axis central to understanding CKD progression [117]. Among the most studied gut microbiota-derived metabolites are SCFAs, uremic toxins such as indoxyl sulfate and p-cresyl sulfate, and TMAO (Figure 3) [118, 119].
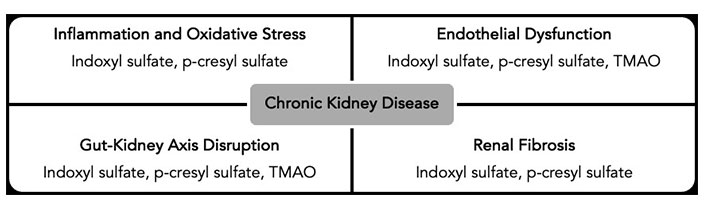
Mechanisms of gut microbiota-derived metabolites in CKD progression. TMAO: trimethylamine-N-oxide; CKD: chronic kidney disease
SCFAs, as previously noted, possess anti-inflammatory and immunomodulatory properties crucial for maintaining gut barrier integrity and supporting systemic immune balance. In the context of CKD, SCFAs may offer protective effects by mitigating inflammation and oxidative stress, which are implicated in CKD progression [120, 121]. Conversely, uremic toxins such as indoxyl sulfate and p-cresyl sulfate, byproducts of gut microbial metabolism of amino acids like tryptophan and tyrosine, accumulate in the bloodstream due to impaired renal clearance in CKD. These toxins contribute to kidney fibrosis by promoting the proliferation of renal interstitial fibroblasts and enhancing extracellular matrix deposition [122, 123]. They also drive inflammation and vascular damage, thereby worsening CKD [121, 124–126]. As mentioned earlier, TMAO is linked to increased cardiovascular risk, a major cause of mortality in CKD patients [127]. TMAO promotes atherosclerosis and vascular calcification, further aggravating CKD and its related complications [127].
Understanding the role of gut microbiota-derived metabolites in CKD opens new avenues for therapeutic intervention. Increasing the intake of dietary fibers can enhance SCFA production, which may protect against CKD progression by reducing inflammation and oxidative stress [128]. Probiotics and prebiotics, by restoring a healthy gut microbiota, may help reduce the production of harmful metabolites [129]. Additionally, oral adsorbents that bind uremic toxins in the gut can reduce their systemic absorption, thereby lowering their plasma concentrations and mitigating their harmful effects on the kidneys [130]. Targeting specific metabolic pathways involved in the production of uremic toxins and TMAO could also provide novel therapeutic approaches to slow CKD progression [127]. Future research should focus on further elucidating the gut-kidney axis and developing targeted interventions to harness the therapeutic potential of gut microbiota modulation.
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease characterized by the production of autoantibodies against nuclear antigens, leading to widespread inflammation and tissue damage. The pathogenesis of SLE involves complex interactions between genetic, environmental, and immunological factors, with emerging evidence highlighting the role of the gut microbiota in disease progression [131].
In patients with SLE, dysbiosis is often observed, with a reduction in butyrate-producing bacteria such as Faecalibacterium prausnitzii and an increase in pathogenic bacteria such as Ruminococcus gnavus [132]. This imbalance contributes to the pro-inflammatory state, worsening disease symptoms. For example, the decrease in butyrate-producing bacteria disrupts immune regulation, reducing differentiation of Tregs and increased production of pro-inflammatory cytokines such as IL-6 and TNF-α [133].
Additionally, tryptophan metabolism by gut bacteria influences the immune response in SLE. The kynurenine pathway, a major route of tryptophan catabolism, generates metabolites such as kynurenine and quinolinic acid, which can modulate T-cell responses. Elevated levels of these metabolites have been associated with disease activity in SLE, suggesting a link between tryptophan metabolism and autoimmunity [134]. The combined effects of gut dysbiosis, altered immune responses, and disrupted tryptophan metabolism underscore the critical role of the gut-immune axis in SLE and highlight the need for further exploration of microbiota-targeted therapies to modulate disease progression.
Multiple sclerosis (MS) is an autoimmune disorder characterized by demyelination of neurons in the central nervous system (CNS), resulting in a variety of neurological symptoms. The gut-brain axis, a bidirectional communication network between the gut microbiota and the CNS, has been increasingly recognized for its significance in the development and progression of MS.
Gut microbiota-derived SCFAs, particularly propionate, have been shown to protect against CNS autoimmunity by promoting the differentiation of Tregs and enhancing the integrity of the blood-brain barrier (BBB) [135]. In patients with MS, dysbiosis is commonly observed, marked by a reduction in SCFA-producing bacteria and an increase in pro-inflammatory species such as Akkermansia muciniphila [136]. This imbalance may contribute to increased gut permeability and systemic inflammation, both of which are implicated in the pathogenesis of MS.
Furthermore, BA metabolism by the gut microbiota has been implicated in MS. BAs can modulate immune responses by activating of receptors such as FXR and the G protein-coupled bile acid receptor 1 (GPBAR1). Alterations in BA metabolism have been observed in MS patients, including reduced levels of anti-inflammatory BA like lithocholic acid, which may contribute to disease progression and severity [136, 137]. The disruption in both SCFA and BA metabolism highlights the importance of gut microbiota in influencing immune responses and CNS inflammation in MS.
Psoriasis is a chronic inflammatory skin disease characterized by hyperproliferation of keratinocytes and infiltration of immune cells into the skin. While the exact cause of psoriasis is not fully understood, gut microbiota-derived metabolites are thought to play a role in modulating immune responses and skin inflammation.
In patients with psoriasis, dysbiosis is commonly observed, characterized by a reduction in butyrate-producing bacteria, which may contribute to the chronic inflammatory state that defines the disease [138]. Butyrate, a SCFA, plays a key role in immune regulation, and its deficiency disrupts immune homeostasis, potentially worsening inflammation. Altered tryptophan metabolism has been linked to psoriasis, with elevated levels of pro-inflammatory metabolites from the kynurenine pathway, such as 3-hydroxykynurenine, thought to drive the Th17-mediated immune responses that are characteristic of psoriasis [139]. Moreover, BAs also play a role in maintaining skin health by promoting skin barrier integrity and modulating immune function. In psoriasis, dysbiosis leads to changes in BA metabolism, resulting in increased levels of pro-inflammatory secondary BAs, which may further exacerbate skin inflammation. The intricate interplay between gut microbiota, tryptophan metabolism, and BA dysregulation highlights the critical role of the gut-skin axis in the pathogenesis of psoriasis, highlighting potential therapeutic targets in modulating these pathways to alleviate disease symptoms [140].
Rheumatoid arthritis (RA) is a chronic autoimmune disease characterized by inflammation of the joints, leading to pain, swelling, and eventual destruction. The role of the gut microbiota in RA has garnered significant attention, with dysbiosis thought to play a key role in the initiation and progression of the disease.
In patients with RA, the reduced presence of SCFA-producing bacteria, such as those responsible for generating butyrate and propionate, significantly effects on immune regulation. This decline impacts the immune system by impairing the formation of Tregs and allowing for an increase in pro-inflammatory cytokines like IL-1β and TNF-α, both of which are key contributors to the inflammatory cascade in RA [22]. Tryptophan metabolism is also altered in RA, with higher levels of pro-inflammatory metabolites from the kynurenine pathway exacerbating the disease by influencing T cell activity [141]. Additionally, altered BA metabolism has been linked to disease activity, as changes in BA profiles further contribute to immune disruption and inflammation [142]. This complex interaction between SCFAs, tryptophan metabolites, and BA dysregulation highlights the significant role of the gut microbiota in driving RA pathology.
Asthma is a chronic respiratory condition affecting more than 300 million people globally, with various clinical phenotypes, primarily categorized into Th2-high and Th2-low endotypes [143]. The most common form is driven by a Th2 immune response, which leads to the overproduction of pro-inflammatory cytokines such as IL-4, IL-5, and IL-13. Microbial metabolites, including SCFAs, tryptophan derivatives, polyamines, and p-cresol sulfate, have been shown to modulate immune responses by regulating T cells and dendritic cells, thereby contributing to the attenuation of asthma symptoms through anti-inflammatory mechanisms [144].
SCFAs play a protective role against allergic asthma. Studies have shown that SCFA levels in children are inversely correlated with asthma risk, and experimental models demonstrate that SCFAs can reduce airway inflammation [145]. Tryptophan metabolism also influences immune responses in asthma, with microbial derivatives such as D-tryptophan and kynurenine reducing airway hyperresponsiveness and downregulating Th2-mediated inflammation [146]. Moreover, polyamines and metabolites like p-cresol sulfate further emphasize the influence of microbial products on lung function and inflammation [18]. Spermidine, a polyamine, has been found to reduce the severity of asthma and prevent tissue damage. At the same, p-cresol sulfate, a product of L-tyrosine metabolism, limits dendritic cell recruitment in the lungs, thereby minimizing inflammation [147]. These interactions between the microbiome and the immune system suggest that modulating microbial metabolites could offer promising therapeutic strategies for managing asthma.
Atopic dermatitis (AD) is a chronic inflammatory skin disorder influenced by genetic, environmental, and immune factors. It is particularly prevalent in young children, making it one of the most common pediatric skin conditions globally. Recent data show an increasing prevalence of AD, now affecting roughly 20% of children and 10% of adults, underscoring its significance as a widespread and concerning dermatological issue [148, 149].
The skin’s microbial communities are critical in maintaining its protective barrier and immune balance. In healthy skin, commensal bacteria like Staphylococcus epidermidis and Staphylococcus hominis produce antimicrobial peptides that inhibit pathogens such as Staphylococcus aureus. However, in AD, an overgrowth of Staphylococcus aureus disrupts this balance, leading to decreased microbial diversity, which has been inversely correlated with the severity of the disease. This imbalance contributes to a compromised skin barrier, exacerbating inflammation and making the skin more susceptible to infections [150].
AD patients also exhibit reduced levels of anaerobic bacteria that produce SCFAs, such as Bifidobacterium and Faecalibacterium prausnitzii [151]. These SCFAs are essential for lowering skin pH and protecting against infection and inflammation [152]. The decrease in SCFA-producing bacteria leads to skin and gut microbiome disturbances, potentially contributing to the inflammatory processes in AD. Studies have linked lower SCFA levels in early childhood with an increased risk of developing AD and other allergic conditions [153].
Additionally, lower levels of tryptophan-derived metabolites have been observed in AD patients, further affecting skin barrier function. Microbial metabolites like indole-3-aldehyde (IAlD) can alleviate AD symptoms by binding to the AhR in skin cells, enhancing barrier integrity, and modulating immune responses [154]. These disruptions in the skin microbiota and the decline in protective microbial metabolites highlight the integral role of the gut-skin axis in the pathogenesis of AD.
The relationship between gut microbiota-derived metabolites and the host immune system opens promising avenues for novel therapeutic strategies in disease management. Microbial metabolites, such as SCFAs, BAs, microbial-derived vitamins, and tryptophan catabolites, play important roles in regulating immune responses, reflecting the diversity and complexity of the gut microbiome itself [37]. Emerging evidence suggests that modulating these metabolites could provide innovative approaches to managing inflammatory diseases effectively [155].
An integrative approach, such as Molecular Pathology Epidemiology (MPE), provides a powerful framework for exploring the connections between the gut microbiome, environmental factors, immunity, and disease. MPE merges molecular biology, pathology, and epidemiology to investigate how environmental exposures influence molecular changes that impact clinical outcomes [156]. By identifying molecular biomarkers, MPE helps elucidate the mechanisms through which environmental factors alter the gut microbiome and immune system, influencing disease progression and therapeutic outcomes [157]. This interdisciplinary approach has the potential to deepen our understanding of disease mechanisms and promote personalized prevention strategies, ultimately improving patient care.
Future research should explore the ways in which microbial metabolites influence immune cell function, focusing on their potential as therapeutic tools. By investigating how specific microbial strains and their byproducts shape immune functions like differentiation and activation, researchers can uncover innovative methods to restore balance in disrupted microbiomes. Translating these findings into clinical practice could revolutionize both the diagnosis and treatment of such conditions. Personalized therapies, guided by detailed profiles of an individual’s microbiome and its metabolic output, promise targeted interventions that address the unique needs of each patient. These efforts will deepen our understanding of the microbiome’s role in health and disease, driving more precise and effective care.
The intricate interplay between the gut microbiota and host health highlights the potential of microbiota-targeted therapies in managing chronic inflammatory diseases. As research continues to unravel the complex relationships between diet, microbiota, and disease, the prospect of modulating the gut microbiota to enhance health and treat disease becomes increasingly viable. The future of treating chronic inflammatory conditions lies in leveraging these relationships, offering hope for innovative, effective, and personalized therapeutic strategies. This integrated approach promises not only to improve disease management but also to enhance overall well-being, ushering in a new era of healthcare where gut microbiota plays a central role in maintaining human health.
AD: atopic dermatitis
AhR: aryl hydrocarbon receptor
BAs: bile acids
BCAAs: branched-chain amino acids
BGA: brain-gut axis
CAD: coronary artery disease
CD: Crohn’s disease
CKD: chronic kidney disease
CNS: central nervous system
CVDs: cardiovascular diseases
FXR: farnesoid X receptor
GI: gastrointestinal
GLP-1: glucagon-like peptide-1
GPRs: G-protein-coupled receptors
HDAC: histone deacetylase
IBD: inflammatory bowel disease
IBS: irritable bowel syndrome
IL-22: interleukin-22
LPS: lipopolysaccharides
MASLD: metabolic dysfunction-associated steatotic liver disease
MPE: Molecular Pathology Epidemiology
MS: multiple sclerosis
NF-κB: nuclear factor kappa-B
NLRs: nucleotide-binding oligomerization domain-like receptors
PYY: peptide YY
RA: rheumatoid arthritis
RS: resistant starch
SCFAs: short-chain fatty acids
SLE: systemic lupus erythematosus
T2DM: type 2 diabetes mellitus
TGR5: Takeda G-protein-coupled receptor 5
TLRs: Toll-like receptors
TMA: trimethylamine
TMAO: trimethylamine-N-oxide
TNF-α: tumor necrosis factor-α
Tregs: regulatory T cells
UC: ulcerative colitis
AV: Conceptualization, Data curation, Formal analysis, Writing—original draft, Writing—review & editing. BLR: Writing—review & editing. KH: Data curation, Writing—original draft, Writing—review & editing. ARVS, MS, and SD: Data curation, Formal analysis, Writing—original draft, Writing—review & editing. DAJ: Conceptualization, Supervision, Writing—original draft, Writing—review & editing.
All authors declare no conflict of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
© The Author(s) 2025.
Open Exploration maintains a neutral stance on jurisdictional claims in published institutional affiliations and maps. All opinions expressed in this article are the personal views of the author(s) and do not represent the stance of the editorial team or the publisher.
Copyright: © The Author(s) 2025. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Olha Denefil ... Khrystyna Loza
Weinan Du ... Juxiu Li
Shin Takasawa ... Maiko Takeda
Zeneng Wang ... Robert Koeth
