Affiliation:
1Department of Biochemistry, Nara Medical University, Kashihara 634-8521, Japan
Email: shintksw@naramed-u.ac.jp
ORCID: https://orcid.org/0000-0002-4066-0199
Affiliation:
2Department of Diagnostic Pathology, Nara Medical University, Kashihara 634-8522, Japan
Affiliation:
2Department of Diagnostic Pathology, Nara Medical University, Kashihara 634-8522, Japan
Explor Med. 2025;6:1001278 DOI: https://doi.org/10.37349/emed.2025.1001278
Received: June 12, 2024 Accepted: December 21, 2024 Published: February 05, 2025
Academic Editor: Zeneng Wang, Cleveland Clinic, USA
The article belongs to the special issue Gut Microbiota Derived Metabolites and Chronic Inflammatory Diseases
Regenerating gene (Reg) was first isolated in 1988 and proposed to be specifically expressed in rat regenerating pancreatic islets. Since then, many genes homologous to Reg have been discovered in other species, including humans, mice, hamsters, rabbits, sheep, dogs, cats, pigs, giant pandas, chickens, and frogs. Moreover, Reg and its related genes (Reg family genes) have been classified as types I, II, III, and IV. They are closely associated with cell and tissue regeneration, cell proliferation, and anti-apoptosis in various tissues and cells including pancreatic β cells. In particular, focusing on the digestive organs and tissues, there have been reports that they play important roles not only in stomach, colon, liver, and pancreatic duct cancer, but also in intestinal epithelial cells, especially inflammatory bowel diseases (IBD), such as Crohn’s disease (CD) and ulcerative colitis (UC). This review describes and discusses the expression of Reg family genes in intestinal epithelial cells of those affected by IBD and the molecular mechanisms underlying this expression.
The lack or deficiency of functional insulin-producing pancreatic β cells has led to the development of diabetes. Replicating β cells are estimated to comprise only approximately 1% of the total β cells in the islets of Langerhans. Over 30 years of research into pancreatic β cell growth and regeneration research has shown that several growth factors act as mitogens that stimulate β cell proliferation [1]. Ninety percent of pancreatectomy rats had positive urine glucose excretion one to three months after surgery. The islets of Langerhans in the remaining pancreas of these rats were rare, small, and frequently showed fibrotic degeneration and degranulation [2, 3]. However, Yonemura et al. [4] showed that 90% of pancreatectomized rats treated daily with an intraperitoneal injection of poly(ADP-ribose) polymerase (PARP; EC 2.4.2.30) inhibitors, such as nicotinamide (pyridine-3-carboxamide) or 3-aminobenzamide, did not develop diabetes. Urinary glucose excretion levels were significantly reduced in rats treated with PARP inhibitors injection. The islets of Langerhans in the PARP inhibitor-injected rats were significantly larger than those in the control group [4]. The number of islets per square centimeter in 90% of pancreatectomized rats receiving PARP inhibitors was also larger than in rats without daily injections of PARP inhibitors [4]. Immunohistochemical staining of the remaining pancreas revealed that nearly the entire area of the enlarged islets in rats treated with PARP inhibitors was densely stained for insulin (β cells). On the other hand, glucagon-positive cells (α cells) were localized to the periphery of the enlarged islets of Langerhans in the remaining pancreas. Their quantity and localization of α cells in the enlarged islets were almost identical to those in normal rat pancreatic islets. These immunohistochemical findings indicate that the β cell population that was increased in the remaining pancreatic islets of rats treated with PARP inhibitors.
Terazono et al. [5] isolated regenerating islets and constructed a λgt10 cDNA library from rat regenerating islets for searching mRNA(s) specifically expressed in regenerating islets. They found a novel gene encoding a 165 amino acid protein with a signal peptide sequence using differential hybridization of the cDNA library. This new gene was named regeneration gene (Reg) because it could contribute to pancreatic islet regeneration [5]. Terazono et al. [5–9] next isolated human counterpart from the human pancreas cDNA library and found human REG cDNA. The human REG cDNA showed a high degree of homology to the rat Reg (75% and 68% homology in nucleotides and amino acids, respectively). Reg gene expression was found in the regenerating pancreatic islets, but not in normal islets, livers, insulinomas induced by combined administration of streptozotocin (2-deoxy-2{methyl(nitroso)amino]carbonyl}amino)-β-D-glucopyranose) and nicotinamide, or regenerating liver induced by partial hepatectomy. Reg expression during islet regeneration in 90% partially pancreatectomized rats treated with nicotinamide increases one month after partial pancreatectomy, peaks at three months, and reaches nearly undetectable levels one year after surgery. The Reg gene was also expressed in the hyperplastic pancreatic islets of aurothioglucose (gold[I] [2S,3S,4R,5S]-3,4,5-trihydroxy-6-[hydroxymethyl]-oxane-2-thiolate)-injected type 2 diabetes models of non-obese non-diabetic (NON) mice [5]. Reg gene expression observed in both rat regenerating and NON mouse hyperplastic islets strongly suggests an involvement of the Reg gene in pancreatic β cell replication, growth, and maturation.
Watanabe et al. [6] expressed recombinant rat Reg protein in a yeast expression system and purified it. When they administered the rat Reg protein (1 mg/kg/body weight per day) intraperitoneally to rats with 90% partial pancreatectomy, they observed an increase in the incorporation of [3H]thymidine into insulin-producing islet cells and frequent mitoses in the remaining pancreatic islets of Langerhans [10]. On postoperative days 30 and 60, fasting plasma glucose levels in 90% pancreatectomized rats injected daily with the recombinant Reg protein were significantly lower than those in 90% pancreatectomized control rats. After two months, nearly all islets of Langerhans in control rats with 90% partial resection of the pancreas without Reg protein administration were reduced, shrunken, and had irregular contours; fibrous degeneration and degranulation frequently occurred [4]. These observations are consistent with others in pancreatectomized animals [2].
In contrast, the islets in rats treated with recombinant Reg protein were enlarged, and nearly all islets stained positive for insulin [10]. In addition, the administration of recombinant human REG (REG Iα) protein also increased β cell mass and improved diabetes in nonobese diabetic (NOD) mice, a typical mouse model of spontaneous type 1 diabetes [11]. Furthermore, human recombinant REG protein stimulates the uptake of 5’-bromo-2’-deoxyuridine (BrdU) into pancreatic β cells in a concentration-dependent manner in vitro. Anti-REG autoantibodies were detected in some diabetic patients’ serum, and the REG protein-induced incorporation of BrdU into β cells was significantly attenuated by adding diabetic patient serum containing anti-REG autoantibodies [12]. These results strongly suggest that the Reg protein increases replication, regeneration, and/or growth of pancreatic β cells, thereby alleviating diabetes.
Unno et al. [13] constructed DNA that was ligated three copies of the rat insulin II promoter-mouse Reg I gene tandemly and microinjected the DNA into male pronuclei of fertilized mouse eggs to produce transgenic (Tg) mice. The [3H]thymidine incorporation in the insulin II promoter-Reg I Tg mouse islets from both males and females was significantly higher than that of the non-Tg control mouse islets [13]. The insulin II promoter-Reg I Tg mice were outcrossed to NOD female mice, and the resultant F1 male was backcrossed into NOD mice. Insulin II promoter-Reg I Tg-NOD and control NOD mice were generated by intercrossing nine backcrossings. The female insulin II promoter-Reg I Tg-NOD mice showed a significantly delayed onset of diabetes, ascertained based on at least two consecutive determinations of glucosuria compared to control female NOD mice without insulin II-Reg I Tg [13]. They also produced Reg I knockout (KO) mice by conventional homologous recombination in embryonic stem (ES) cells, and the pancreatic islets of KO mice showed a reduced [3H]thymidine incorporation in the islet cell nuclei [13]. When hyperplastic islets were induced by aurothioglucose (0.5 g/kg of body weight) injections in the KO and control mice to induce Reg I expression and hyperplastic islets, the islet volume, estimated by histochemical specimens, of the Reg I KO mice was significantly smaller than that of the aurothioglucose-injected control mice [13], indicating an essential role of endogenous Reg I in maintaining/increasing islet β cells. Liu and his colleagues produced Reg II KO mice. They found that the aging Reg II KO mice (13- to 14-month-old Reg II KO) developed glucose intolerance (high glucose levels in serum were observed at 30, 60, and 120 min after glucose injection) associated with decreased islet β cell ratio (the quantification of the percent of the islet area to total tissue showed a significant 60% decrease in Reg II KO mice) and serum insulin [14].
As mentioned above, Reg proteins may function as autocrine or paracrine growth factors in pancreatic β cells. Because Reg protein is a secreted protein, it has been thought that a cell surface receptor in pancreatic islets must receive its proliferation signals. Its actual nature remains unknown. Kobayashi et al. [15] screened a λZAP II rat islet cDNA expression library and isolated Reg protein-binding protein (receptor) cDNA by binding to the 125I-rat Reg protein. The cDNA contained an open reading frame of 2,760-base pairs encoding a 919-amino acid type II transmembrane protein [with a long extracellular domain (residues 52–919; 868 amino acid residues), a transmembrane domain (residues 29–51), and a short intracellular region at N terminus (residues 1–28)] with apparent molecular weight of 105,000, and the COS-7 cells into which the cDNA (cDNA in pCIneo mammalian expression vector) had been transfected and expressed bound the Reg protein with high affinity (Kd = 4.41 nM) [15]. Introducing the Reg protein receptor cDNA into rat RINm5F β cells significantly increased BrdU incorporation in the transformants and cell numbers in response to the Reg protein [15]. The primary structure of the rat cDNA was highly homologous to human multiple exostoses (EXT)-like genes—especially the human EXT-like gene 3 (EXTL3)/EXT-related gene 1 (EXTR1)—with an amino acid identity of over 97%, indicating that the 919-amino acid Reg protein receptor is encoded by the EXTL3/EXTR1 gene and mediates Reg protein proliferation signals for β cell regeneration. After the report by Kobayashi et al. [15] that the Extl3/Extl1 gene encodes a Reg receptor, Acquatella-Tran Van Ba et al. [16] reported that Extl3/Extl1 is a Reg receptor. Furthermore, several groups have also reported Extl3/Extl1 as a Reg protein receptor [8, 9]. In later studies, additional discoveries about the Reg receptor were reported. The addition of Reg protein in the culture medium of rat RINm5F β cells resulted in the activation (phosphorylation) of transcription activating transcription factor 2 (ATF-2) [17]. β cell-specific disruption for the Reg receptor (Extl3) showed abnormal islet morphology accompanied by reduced β cell replication/proliferation observed by proliferating cell nuclear antigen (PCNA) positive β cells after one week of age [18].
Reg receptor (Extl3) was expressed not only in pancreatic islets of Langerhans but in other cells and tissues, such as normal liver, heart, kidney, spleen, thymus, stomach, small intestine, colon, pancreatic acinar and ductal cells, adrenal gland, pituitary gland, testis, and brain [15, 16]. This suggested the involvement of the Reg-Reg receptor signaling system for cell proliferation in a wide variety of cell types other than pancreatic β cells, such as epithelial cells in the small intestine and colon. Because Reg was found in the regenerating islets of Langerhans, Reg tends to be expressed in a limited number of cells under certain conditions. In contrast, Reg protein receptors were found to be expressed in a wide variety of cells. The Reg-Reg receptor signaling system was considered to be dependent on the expression of the Reg gene(s), but not the Reg receptor.
Many Reg and Reg-related genes have been isolated (see Figure 1), and Reg and Reg-related genes constitute a multigene family, the Reg family [7–9, 19]. Based on the amino acid sequence of the Reg family proteins, the members of the Reg family were grouped into four subclasses: types I, II, III, and IV [7–9] (Figure 1). Recently, chimpanzee Reg Iα (XP_001163554.2), Reg Iβ (XP_001163951.4), Reg IIIα (XP_003816656.1), Reg IIIγ (XP_003816661.1) and Reg IV (XP_016780102.1), western lowland gorilla Reg Iα (XP_004029557.1), Reg IIIα (XP_055234895.1), Reg IIIγ (XP_055234615.1) and Reg IV (XP_055211509.1), Francois’s langur Reg Iα (XP_033031150.1), Reg Iβ (XP_033031147.1), Reg IIIα (XP_033032683.1) and Reg IV (XP_033053315.1), Sumatran orangutan Reg Iα (XP_002811895.2), Reg Iβ (XP_002811894.1), Reg IIIγ (XP_054332180.1) and Reg IV (XP_002810393.2), northern white-cheeked gibbon Reg Iα (XP_003282186.1), Reg Iβ (XP_003282184.2), Reg IIIα (XP_003268777.1) and Reg IV (XP_003268113.1), siamang Reg Iα (XP_055098489.1), Reg Iβ (XP_055097639.1) and Reg IIIγ (XP_055098377.1), pig-tailed macaque Reg Iα (XP_011712048.1), Reg IIIγ (XP_011712043.1), Reg IIIα (XP_011712050.1) and Reg IV (XP_011735163.1), silvery gibbon Reg Iα (XP_032034470.1), Reg Iβ (XP_032034469.1), Reg IIIα (XP_032033947.1) and Reg IIIγ (XP_032033085.1), golden snub-nosed monkey Reg Iα (XP_010378205.1), Reg IIIα (XP_010382870.1) and Reg IV (XP_010358951.1), cotton-top tamarin Reg Iα (KAK2092743.1), Reg Iβ (KAK2092742.1), Reg IIIα (KAK2092744.1) and Reg IIIγ (KAK2092740.1), crab-eating macaque Reg Iα (XP_005575526.1) and Reg Iβ (XP_005575530.1), green monkey Reg Iα (XP_007968256.1), Reg Iβ (XP_007968260.1), Reg IIIα (XP_007968253.1) and Reg IIIγ (XP_007968263.2), white-tufted-ear marmoset Reg Iα (XP_002757620.1), Reg Iβ (XP_002757622.1), Reg IIIγ (XP_035128030.1) and Reg IV (XP_017821669.3), sooty mangabey Reg Iα (XP_011941399.1), Reg IIIα (XP_011941401.1), Reg IIIγ (XP_011941395.1) and Reg IV (XP_011945315.1), Rhesus monkey Reg Iα (XP_014968213.1), Reg Iβ (NP_001181497.1), Reg IIIγ (XP_001112492.1) and Reg IV (XP_001084012.1), crab-eating macaque Reg Iα (XP_005575526.2) and Reg IIIα (XP_005575524.1), gelada Reg Iα (XP_025209412.1), Reg IIIα (XP_025210394.1), Reg IIIγ (XP_025210180.1) and Reg IV (XP_025244087.1), olive baboon Reg Iα (XP_021780704.1), Reg Iβ (XP_021780701.2), Reg IIIα (XP_021780706.1) and Reg IV (XP_021778575.2), Ma’s night monkey Reg Iα (XP_012307533.1), Reg Iβ (XP_012307535.1), Reg IIIγ (XP_012307641.1) and Reg IV (XP_021529810.1), tufted capuchin Reg Iα (XP_032099367.1), Reg Iβ (XP_032099369.1), Reg IIIα (XP_032099373.1), Reg IIIγ (XP_032099366.1) and Reg IV (XP_032127887.1), Panamanian white-faced capuchin Reg Iα (XP_017392173.1), Reg Iβ (XP_017392177.1), Reg IIIα (XP_017392178.1), Reg IIIγ (XP_017392170.1) and Reg IV (XP_017375266.1), Philippine tarsier Reg Iα (XP_008061466.1), Reg Iβ (XP_008061465.1) and Reg IV (XP_008062946.1), Bolivian squirrel monkey Reg Iα (XP_003922600.1), Reg Iβ (XP_003922602.1) and Reg IV (XP_039316867.1), Ugandan red Colobus Reg Iα (XP_026312216.1), Reg Iβ (XP_023043790.1), Reg IIIα (XP_023043801.1) and Reg IV (XP_023078498.1), slow loris Reg Iβ (XP_053445874.1), black snub-nosed monkey Reg Iβ (XP_017708014.1), Reg IIIα (XP_017739229.1) and Reg IV (XP_017719648.1), small-eared galago Reg Iβ (XP_012667807.1), gray mouse lemur Reg Iα (XP_012590838.1), Reg Iβ (XP_012590831.1), Reg IIIγ (XP_045405903.1) and Reg IV (XP_012617318.1), Cape golden mole Reg Iβ (XP_006873819.1), aardvark Reg IV (XP_007946661.1), bear Reg IIIβ (XP_026373137.1) and Reg IV (XP_008704575.1), takin Reg IV (XP_052494122.1), hamster Reg IIIβ (XM_021231969), Reg IIIγ (NM_001281579.1) and Reg IV (XM_021235470.2), squirrel Reg IV (XP_047389427.1), oryx Reg IV (XP_040086853.1), shrew Reg IIIα (XM_006169192.2), Reg IIIγ (XM_006169191.2) and Reg IV (XM_006166291.1), rabbit Reg IIIγ (XM_002709697.2) and Reg IV (XM_002715670.3), American pika Reg IV (XP_058516894.1), canine Reg IIIα (NM_001002945.3) and Reg IV (XM_038499361.1), dingo Reg IV (XP_025309107.1), raccoon dog Reg IIIβ (NP_001002945.2), Reg IIIγ (XP_055168716.1) and Reg IV (XP_055168118.1), red fox Reg IIIγ (XP_025850460.1) and Reg IV (XP_025844996.1), Cavia porcellus (guinea pig) Reg IIIβ (XM_003468964.1) and Reg IIIγ (XM_003468904.4), Bos taurus (bovine) Reg IIIα (NM_205787.1) and Reg IV (NM_001076986.1), Sus scrofa (porcine) Reg IIIγ (XM_005662419.3) and Reg IV (NM_001190251.1), Equus caballus (equine) Reg IIIγ (XM_001498171.6), Felis catus (ferine) Reg IV (XM_023259045.1), bobcat Reg IV (XP_046938644.1), sheep Reg IIIβ (NM_001038014.1), American wolverine Reg IV (KAI5761890.1), goat Reg IV (XP_005677839.1), deer Reg IV (XP_055259912.1), hippopotamus Reg IV (XP_057571303.1), Malayan pangolin Reg IV (XP_017505537.1), otter Reg IV (XP_047582626.1), beaver Reg IV (XP_020043589.1), giant pangolin Reg IV (KAK2495942.1), armadillo Reg IV (XP_012379915.1), giant panda Reg IV (XP_002928439.1), seal Reg IIIβ (XP_021554843.2) and Reg IIIγ (XP_006727443.1), sea lion Reg IIIβ (XP_027478978.1), walrus Reg IIIβ (XP_004411644.1), kangaroo Reg IIIβ (XM_013031828.1), Tasmanian devil Reg IV (XP_003769898.1), Suncus murinus (Asian house shrew) Reg IIIα (LC606672.1) and Reg IV (XP_049621912.1), Rhinolophus ferrumequinum (greater horseshoe bat) Reg IIIα (XM_033125205.1), Desmodus rotundus (common vampire bat) Reg IIIγ (XM_024558600.1) and Reg IV (XM_024570607.1), Gallus gallus domesticus (chicken) Reg IV (NM_001277527.1), Cotumix japonica (Japanese quail) Reg IV (XM_015869386.1) and Xenopus tropicalis (tropical clawed frog) Reg Iβ (XM_002934582.4) and Reg IV (XM_004915988.3) were isolated.
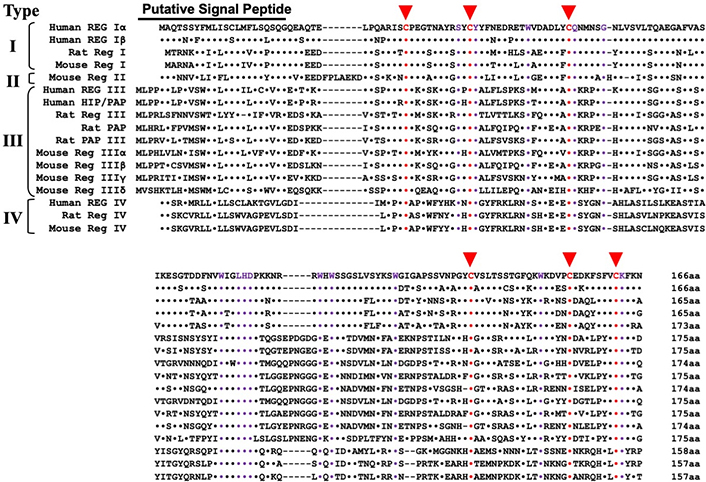
Alignment of primary structures of the Reg gene family. Based on the amino acid sequences (adapted from [7–9, 19, 70]), the Reg family is classified into four subclasses: types I, II, III, and IV. Dots indicate amino acids identical to human REG Iα in the alignment. Dashes indicate gaps in maximal alignment. Six conserved cysteines (C) in the mature proteins are depicted in red. Amino acid residues common to all the Reg family proteins other than the six cysteines are indicated by purple. Types I, II, and III Reg genes are clustered in a restricted region (~100 kbp) of the same chromosome [19, 33, 70]. The human REG-related sequence (RS) is omitted from the alignment because human RS is a pseudogene [6]. Reg: regenerating gene
Types I and II Reg proteins are expressed in islet β cell regeneration [5, 13, 14]. However, type I Reg has been reported to be expressed in pancreatic β cells and human colorectal cancer, gastric mucosa, and enterochromaffin cells of water-immersion restraint rats [7–9]. Type III Reg proteins [pancreatitis-associated proteins (PAPs)] have been involved in the cellular proliferation of intestinal Paneth granule cells, hepatocellular carcinoma, pancreatic exocrine acinar cells, keratinocytes, and Schwann cells (neurolemmocytes); Reg family proteins have been implicated to function not only in pancreatic β cell regeneration. It has been suggested that Reg family protein(s) may be involved in the regeneration/anti-apoptosis of some other cells and tissues than that of pancreatic β cells [7–9]. Bréchot and his colleagues [20] produced mouse albumin promoter-HIP/PAP (human counterpart of Reg IIIβ) Tg mice. They found that liver regeneration after partial (70%) hepatectomy was accelerated in the albumin-HIP/PAP Tg mice.
They also produced Reg IIIβ KO mice by homologous recombination and found that partial hepatectomy-induced liver regeneration of the KO mice was markedly impaired [21]. However, they did not report morphological and biochemical changes in pancreatic β cells and intestines of the Tg and KO mice. Most recently, Chen et al. [22] generated global KO mice for Reg I–III (Reg I, II, IIIα, IIIβ, IIIγ, and IIIδ) genes using the CRISPR/Cas9 system. The Reg I–III KO mice showed no abnormality in general growth, including pancreas development. However, the Reg I–III disrupted mice led to a reduction in pancreatic parenchymal loss, decreased deposition of collagen, and reduced expression of proinflammatory cytokines, such as interleukin (IL)-6, tumor necrosis factor (TNF)α, and IL-1β in chronic pancreatitis [22]. However, they did not analyze/report abnormalities in pancreatic β cells and intestines of the Reg I–III KO mice.
Reg proteins are induced to express in pancreatic β cells/β cell precursors and function as autocrine/paracrine growth/anti-apoptosis factor(s) through the cell surface Reg protein receptor [7–10, 15]. However, after the Reg protein binds to the receptor, it is uncertain how the intracellular molecular mechanisms (signal transduction mechanisms) of the Reg protein induce β cell regeneration/replication. Reg I was found to increase the phosphorylation of ATF-2, which binds to –57 to –52 of the cyclin D1 promoter [17]. The Reg I/ATF-2-induced cyclin D1 promoter activation was attenuated by phosphoinositide 3-kinase (PI(3)K; EC: 2.7.1.137) inhibitors, such as LY294002 (2-(Morpholin-4-yl)-8-phenyl-4H-1-benzopyran-4-one) and wortmannin (1α-(Methoxymethyl)-3,7,17-trioxo-2-oxa-6,4-(epoxymetheno)androsta-5,8-dien-11α-yl acetate) [17]. In Reg I KO mouse islets, the levels of phosphorylated ATF-2, cyclin D1, and phosphorylated retinoblastoma gene (Rb) product were reduced significantly, indicating that the Reg-Reg receptor system stimulates the PI(3)K/ATF-2/cyclin D1 signaling pathway to induce pancreatic β cell regeneration/anti-apoptosis [17].
It has been shown that high concentrations of the Reg I protein (> 300 nM) induce β cell apoptosis, and the hepatocyte growth factor (HGF) (< 2.5 ng/mL) attenuates the β cell apoptosis [15, 23, 24]. During the characterization of the Reg receptor, high concentrations of the Reg I protein induced apoptosis in pancreatic β cells, while the combined addition of IL-6 and dexamethasone (Dx) prevented a high concentration of Reg I-induced β cell apoptosis. The combined addition of IL-6 (20 ng/L) and Dx (100 nM) also induced HGF expression in β cells, and HGF reduced the high concentration-Reg I-induced β cell apoptosis [15, 23]. Yamaoka et al. [25]. reported that Reg I Tg mice were produced by Reg I cDNA under a glucagon promoter. Therefore, a glucagon-Reg I Tg line, which expresses much more Reg I than other lineages in pancreatic α cells, developed diabetes through apoptosis of pancreatic islet cells.
Numbers of the gene structures related to Reg were isolated from rats, mice, humans, and other species; they constitute a multigene family, the Reg gene family [7–9, 26]. Based on the primary structures of the encoded proteins of Reg and the structure-related genes, they are grouped into four subclasses: types I, II, III, and IV (Figure 1). In humans, four functional REG family genes, i.e., REG Iα [5, 6], REG Iβ [27], HIP [28]/PAP [29], and REG III [19], and a pseudogene [REG-related sequence (RS)] [6] are tandemly ordered in the 95 kbp region of chromosome 2p12 [30]. On the other hand, REG IV is located on a different chromosome, chromosome 1p12–13.1 [31, 32]. In the mouse genome, the chromosomal localization of all the Reg family genes except Reg IV, namely Reg I, Reg II, Reg IIIα, Reg IIIβ, Reg IIIγ, and Reg IIIδ, was located in a contiguous 75 kbp region on chromosome 6C3 (in the order of Reg IIIβ-Reg IIIδ-Reg IIIα-Reg II-Reg I-Reg IIIγ) [22, 33]. The Reg IV (encoding Type IV Reg protein) gene was mapped to chromosome 3F3. Type I and type II Reg proteins are expressed in regenerating islets [5, 13, 26] and are involved in β cell regeneration and neogenesis [10–14, 34–36].
Reg family proteins may be involved in the regeneration of insulin-producing cells and in cellular proliferation and regeneration in pancreatic exocrine acinar cells [37], gastrointestinal epithelial cells [38–45], hepatocytes [20, 21, 46–48], cardiovascular cells [8, 9], salivary ductal epithelial cells [49–51], bone and muscle cells [52, 53], and neuronal cells [16]. Mouse Reg IIIβ (PAP) has been considered as a Schwann cell (neurolemmocytes) mitogen that accompanies the regeneration of motor neurons [54] and functions as a neurotrophic factor for motor neurons [55, 56].
In 2000, a group from the St. James’s University Hospital in Leeds, UK reported that death of early colorectal cancer patients (18 patients in stage I and 60 patients in stage II; T1–2, N0M0) could be predicted using the REG family (REG Iα and HIP/PAP) gene expression [57]. In 2003, Yonemura et al. [58] reported that the REG Iα gene expression, detected using RT-PCR, immunofluorescent microscopy, and immunoblot analysis, was no relation detected between REG Iα mRNA status and serosal invasion, lymph node metastasis, lymphatic invasion, and macroscopic type. However, the REG Iα expression was closely related to the infiltrating property of gastric carcinomas and was a prognostic indicator of gastric differentiated adenocarcinoma. The four-year survival was shown to be significantly better in patients with REG Iα-negative gastric carcinomas than in those with REG Iα-positive carcinomas (91% vs. 51% survival, respectively; P < 0.05). Since then, a correlation between REG family gene expression and cancer patient prognosis has been reported in gastric, esophageal, lung, head and neck, and colorectal carcinomas [59–65]. These observations strongly suggest that the expression of the Reg gene family engages in the proliferation/anti-apoptosis of a wide variety of cell types, including cancer cells.
Crohn’s disease (CD) and ulcerative colitis (UC), two major forms of inflammatory bowel diseases (IBD), are both characterized by chronic and destructive intestinal inflammation of unknown cause(s). Approximately one-third of IBD patients experience persistent irritable bowel syndrome (IBS)-like gastrointestinal symptoms despite the absence of objective evidence of disease activity and patients of IBD with an elevated risk of colorectal cancer(s). Despite advances in understanding the cellular/molecular mechanism(s) underlying IBD in intestinal mucosa over the past decade, the exact pathogenesis of IBD remains undefined.
Overexpression of REG Iα and REG Iβ mRNA has been reported in resected colonic tissues taken from those with CD and UC [66]. We also showed the overexpression of HIP/PAP and REG III in IBD in resected colonic mucosa [38]. Overexpression of REG Iα mRNA and protein in UC, particularly dysplasia or cancer, and the possible role of REG Iα as a marker or exacerbating factor of UC-related tumors have also been reported.
Microorganisms, such as enterobacteria, are involved in intestinal inflammation, including IBD. Still, it remains unclear which genes are expressed in response to luminal microbiota in intestinal epithelial cells and whether these genes are also involved in pathological mucosal inflammation. Germ-free mice were orally challenged with a bacterial suspension (freshly prepared fecal suspension) prepared from conventionally housed specific pathogen-free (SPF) mice (a process known as bacterial reconstitution). Thereafter, Ogawa et al. [38] conducted a differential display analysis to determine which gene expression in gut epithelial cells and found that Reg family genes as candidates. They dissolved dextran sulfate sodium (DSS), a synthetic sulfated branched polysaccharide derivative of dextran with a molecular weight of 36–50 kDa and multiple uses in biomedical and clinical research in drinking water and adjusted it to a concentration of 3%. One important property of DSS is that it triggers colitis in rodents (mice and rats) by binding to medium-chain-length fatty acids in the colon mucosa and inducing intestinal inflammation within 7–10 days.
The mice were kept under SPF conditions and allowed free access to 3% DSS solution as drinking water for 5–10 days. The expression of the genes that had been identified by differential display was examined not only in a bacterial reconstitution model, a DSS-induced colitis model, and human IBD intestinal epithelial cells. Type III Reg family genes, especially Reg IIIβ and Reg IIIγ, were strongly induced in gut epithelial cells following bacterial reconstitution and drinking of DSS solution resulted in the colitis [38]. Recently, Shindo et al. [67] at Toho University produced Reg IIIβ KO mice using the CRISPR-Cas9 system and found that 1.5% DSS drinking-induced colitis, characterized by body weight loss and infiltration of neutrophils, was exacerbated in Reg IIIβ KO mice compared to wild-type control mice.
The mRNA expression of HIP/PAP, a human counterpart of Reg IIIβ (see Figure 1), was increased in intestinal epithelial cells of patients with IBD. Reg IIIβ (PAP) expression was localized in mouse intestinal epithelial cells, including goblet cells and columnar cells. On the other hand, HIP/PAP-expressing cells were correlated with metaplasia of Paneth cells in the human colon. Epithelial expression of Reg IIIα, Reg IIIβ, and REG IIIγ in mice or HIP/PAP in IBD patients was induced under mucosal inflammation caused by exposure to resident bacteria or DSS and inflamed IBD colon [38].
Progression and recovery in IBD depend on the balancing of mucosal damage and repair. Some of Reg family members have been reported to be expressed in CD and UC patients’ intestinal mucosa and to function as proliferative mucosal factors in IBD intestinal mucosa. However, it is still unclear whether all the REG family genes are expressed in IBD intestinal mucosa. Tsuchida et al. [43] analyzed the expression of all the functional REG family genes (REG Iα, REG Iβ, REG III, HIP/PAP, and REG IV) in biopsy specimens from UC and CD patients [88 IBD (49 CD and 39 UC) and 44 controls] using real-time RT-PCR. They found that REG Iα, REG Iβ, and REG IV genes were overexpressed in CD biopsy samples and that the REG IV gene was overexpressed in UC samples. On the other hand, the expression of type III human REG genes (REG III and HIP/PAP) was unchanged in the IBD colon samples compared to controls.
IL-6 and IL-8 induced some REG family genes in colon epithelial cells. Several cytokines/growth factors such as IL-22, TNFα, IL-17A, basic fibroblast growth factor (bFGF), HGF, and epidermal growth factor (EGF), have also been reported to induce REG family mRNA(s) in several tissues and cells. To clarify whether the REG Iα gene is upregulated in response to cytokine(s)/growth factor(s) in intestinal epithelial cells, reporter plasmid(s) containing REG Iα promoter with firefly luciferase (EC 1.13.12.7) were constructed, and transfected them into colon epithelial cells (LS-174T and HT-29); the cells were stimulated by the cytokines/growth factors, and the promoter activities were measured. REG Iα transcription was increased by the addition of IL-6 (20 ng/mL) and/or IL-22 (20 ng/mL) [43]. To clarify the cis-element of REG Iα promoters (–1190 to +26) in response to IL-22 or IL-6, we constructed reporter plasmids fusing various lengths of REG Iα promoters to the luciferase gene, introduced them into human colon epithelial cells (LS-174T and HT-29), and measured the promoter activities. Progressive deletion down from –1190 to –220 in the REG Iα promoter did not alter the IL-22-induced promoter activities. However, additional deletions to –220 to –211, –179 to –156, and –146 to –130 caused significant decreases in the promoter activities, indicating that the regions contain cis-elements essential for the IL-22-induced REG Iα promoter activities.
Each region has a putative transcription factor binding site for myeloid zinc finger 1 (MZF1) (also known as MZF1B, ZFP98, ZNF42, and ZSCAN6), related transcriptional enhancer factor-1 (RTEF1) [also known as TEF-3 or TEA domain transcription factor 4 (TEAD4)], and signal transducer and activator of transcription 3 (STAT3) (also known as ADMIO, APRF, and HIES), respectively [43]. The regions essential for the IL-22-induced REG Iα promoter activation were also responsible for the IL-6-induced REG Iα promoter activation. Therefore, IL-6-induced REG Iα expression occurs in the same mechanism as that of IL-22-induced REG Iα expression. To investigate the significance of MZF1, RTEF1/TEAD4, and STAT3 in IL-22-induced REG Iα expression, we transfected small interfering RNA (siRNA) for MZF1, RTEF1/TEAD4, and STAT3 [43, 44]. Introducing siRNAs abolished the IL-22-induced REG Iα expression [43, 44].
In REG Iβ expression, IL-22 induced REG Iβ promoter activation, but the other cytokines/growth factors, including IL-6, did not. The –274 to –260 region was essential for the IL-22-induced REG Iβ expression, and that the region contains a putative binding site for helicase-like transcription factor (HLTF)/forkhead box N2 (FOXN2). To clarify the essential role of HLTF/FOXN2 for REG Iβ expression, we found that the introduction of HLTF/FOXN2 siRNA attenuated the IL-22-induced REG Iβ mRNA expression (Figure 2) [43, 44].
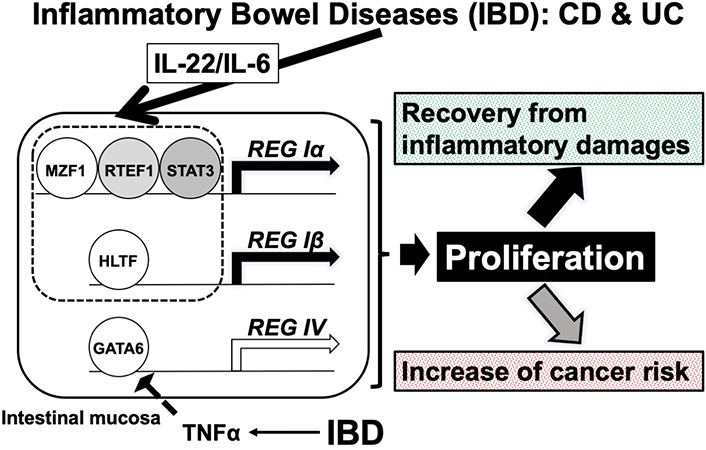
A possible mechanism of REG Iα and REG Iβ gene expression in IBD. The expression of REG Iα and REG Iβ was induced to protect/recover the intestinal epithelial cells from immune-mediated damage via the proliferation of intestinal epithelial cells in IBD. However, it may sometimes result in an increased risk of colorectal cancer significantly. Reduction of REG IV expression in response to TNFα may reduce the risk of colon cancer. MZF1: myeloid zinc finger 1; RTEF1: related transcriptional enhancer factor-1; STAT3: related transcriptional enhancer factor-1; REG: regenerating gene; HLTF: helicase-like transcription factor; GATA: GATA-binding transcription factor; TNF: tumor necrosis factor
In the REG IV promoter (–1053 to +22), no enhancement of promoter activity was detected following the addition of any of cytokine/growth factors (20 ng/mL IL-6, 10 nM IL-8, 1 μg/mL IL-17A, 20 ng/mL IL-22, 20 ng/mL TNFα, 50 ng/mL HGF, 10 nM bFGF, and 10 nM EGF) to LS-174T intestinal epithelial cells. However, REG IV promoter activity was significantly reduced by the addition of TNFα. We assessed the introduction of siRNAs for caudal-type homeobox protein 2 (CDX2), suppressor of cytokine signaling 3 (SOCS3), and GATA-binding factor 6 (GATA6), as CDX2 and GATA6 engaged in REG IV gene expression, and TNFα was reported to regulate SOCS3 expression. GATA6 siRNA introduction abolished the TNFα-induced reduction of REG IV, indicating that GATA6 is an essential transcription factor for REG IV expression [43, 44]. We then searched targeted microRNA (miR) and its possible binding site(s) and found that mRNAs for REG IV and GATA6 have a target sequence for miR-24 and target sequences for miR-143 and miR-363, respectively. The miR-24, -143, and -363 measurements revealed that miR-363 was increased by TNFα treatment [43, 44].
To clarify whether the expression of GATA6 and REG IV in TNFα-treated cells is controlled by miR-363, miR-363 mimic, and miR-363 mimic NC were introduced into LS-174T cells just before TNFα treatment. The TNFα reduced GATA6 and REG IV mRNAs were abolished by the introducing the miR-363 mimic (Figure 2); this indicated that TNFα treatment upregulates miR-363 levels but reduces GATA6 levels. As a result, the REG IV mRNA was significantly reduced in the colorectal cells [44].
Mucosal lesions in the intestine associated with IBD depend on the balance between damage and repair of the mucosal epithelium. Some REG family members (REG Iα, REG Iβ, and REG IV) are expressed in CD and UC epithelium and function as proliferative mucosal factors in CD and/or UC. REG Iα and REG Iβ were induced in the cell culture system by IL-6/IL-22 as described in the previous section. REG IV was also upregulated in CD and UC biopsy samples. However, REG IV upregulation/downregulation using IL-6, IL-22, or TNFα was not observed in the LS-174 and/or HT-29 cell culture system. We found that REG IV mRNA was significantly upregulated by 1 μg/mL lipopolysaccharide (LPS) in both LS-174 and HT-29 human intestinal epithelial cells. However, the other members of the human REG family (REG Iα, REG Iβ, REG III, and HIP/PAP) and a potential LPS receptor, receptor for advanced glycation endproducts (RAGE) were not upregulated by LPS.
To determine whether REG IV is a growth factor/an anti-apoptotic factor in intestinal epithelial cells, we introduced siREG IV RNA into LS-174 and HT-29 cells for knocking-down REG IV and added LPS into the medium. LPS-induced cell proliferation was significantly inhibited by siREG IV-introduced cells, suggesting that REG IV was expressed in intestinal epithelial cells in response to LPS stimulation. Furthermore, it functions as a growth factor/an anti-apoptotic factor for intestinal epithelial cells in the presence of LPS [45]. LPS is a large molecule composed of lipids and polysaccharides. They consist of a covalently bound O antigen of eubacteria, an outer core, and an inner core and are present in the bacterial capsule (outermost membrane of the cell envelope) of gram-negative bacteria such as Escherichia coli (E. coli) and Salmonella enterica (formerly Salmonella choleraesuis). Intestinal inflammation is a host defense response to invading microbial toxins (such as LPS), which are important risk factors for colorectal cancer and IBD.
Disruption of the mucosal barrier and failure of post-disruption regenerative proliferation are the primary mechanisms of intestinal inflammation. Intestinal inflammation thins the mucosal layer, reduces luminal coverage and adhesions, increases epithelial cell apoptosis and permeability, disrupts intestinal integrity by inhibiting cell proliferation, and leads to the loss of barrier function. This can ultimately lead to acute bowel dysfunction. In addition, intestinal mucosal dysfunction leads to the spread of metabolites, endotoxins, and bacteria, and the effects caused by enteric bacteremia exacerbate sepsis and multiple organ dysfunction syndrome. These results could well explain the recent results from Qi et al. [68] that Reg IV KO mice, produced via the CRISPR/Cas9 method, reduced complement bactericidal abilities against E. coli.
To determine whether the LPS-induced increases in REG IV mRNAs in LS-174T and HT-29 intestinal cells were caused by transcriptional activation of the REG IV gene, a human REG IV promoter (–1053 to +22) was fused to the luciferase gene and transfected it into LS-174T and HT-29 human intestinal epithelial cells. After LPS stimulation, REG IV promoter activity was measured. As a result, LPS did not significantly increase the REG IV transcriptional activity in LS-174T or HT-29 human intestinal epithelial cells, suggesting that the upregulation of REG IV in response to LPS stimulation is not regulated by transcription [44, 45]. A possible explanation was that the LPS-induced increase in REG IV expression is controlled post-transcriptionally. Therefore, we searched targeted miR for REG IV mRNA, which revealed that REG IV mRNA has a potential target sequence for miR-24.
We then measured miR-24 levels in LPS-treated and -untreated cells and found that the levels of miR-24 in LPS-treated cells were significantly lower than those in vehicle-treated cells. To confirm whether REG IV expression in LPS-treated cells is controlled by the levels of miR-24, miR-24 mimic and miR-24 mimic NC were introduced into LS-174T and HT-29 intestinal epithelial cells. After LPS treatment, REG IV mRNA in the cells and REG IV protein secreted in the culture medium were significantly increased. Figure 3 shows the LPS-induced increases in REG IV mRNA in the cells; REG IV protein in the culture medium was attenuated by the transfection of miR-24 mimic but not by control RNA (miR-24 mimic NC). These findings confirm that LPS downregulates the miR-24 in human intestinal epithelial cells. As a result, the REG IV mRNA and secreted REG IV protein are increased through the miR-24 mediated mechanism [45].
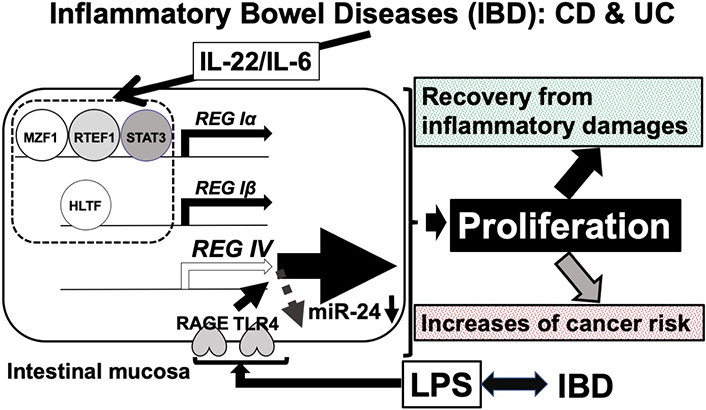
A possible mechanism of REG family (REG Iα, REG Iβ, and REG IV) gene upregulation in IBD. Upregulation of REG Iα, REG Iβ, and REG IV mRNAs may have two opposing features. The expression of REG Iα, REG Iβ, and REG IV could induce protection/recovery of the intestinal epithelium from immune-mediated damages (mainly by IL-22/IL-6) via intestinal epithelial cell proliferation by REG family proteins in IBD. LPS-induced REG IV expression via the reduction of miR-24. The LPS signal was mediated through cell surface RAGE and/or toll-like receptor 4 (TLR4) receptors [45]. In IBD conditions, damaged intestinal epithelial cells were replenished by proliferation. REG IV could induce the replication of intestinal epithelial cells [45]. However, it may sometimes result in a growing risk of colorectal cancer. Therefore, the long-lasting elevation of IL-22/IL-6 in serum and the IL-22/IL-6-induced upregulation of REG family genes in intestinal epithelial cells should be carefully monitored and controlled for healthy recovery of the intestinal epithelium in IBD patients. After the damaged intestinal epithelium is recovered, anti-IL-22/anti-IL-6 antibodies or antibodies against REG family proteins may be used as a new preventive therapy to minimize the risk of colorectal cancer in UC and CD patients. MZF1: myeloid zinc finger 1; RTEF1: related transcriptional enhancer factor-1; STAT3: related transcriptional enhancer factor-1; REG: regenerating gene; HLTF: helicase-like transcription factor; RAGE: receptor for advanced glycation endproducts; LPS: lipopolysaccharide
The expression of Reg family genes in IBD situation and their possible molecular mechanisms have been clarified, as described above. REG protein expression and circulating autoantibodies against the REG protein have also been reported in other autoimmune diseases such as diabetes [12] and Sjögren’s syndrome [49, 51]. Until now, the possible involvement of autoantibodies against REG (Reg family) proteins in IBD and Celiac disease patients has been suggested [40]. Detection and measurement of autoantibodies against REG protein in patient serum has been developed using immunoblot analysis, with some modifications [12, 49, 69]. One microliter of patient serum is sufficient to detect and measure the autoantibodies against REG protein. We anticipate further analyses to detect/measure autoantibodies against the REG family proteins in IBD and immune-mediated intestinal/digestive organ diseases such as Celiac disease, immunodysregulation polyendocrinopathy enteropathy X-linked (IPEX) syndrome, autoimmune gastrointestinal dysmotility (AGID), primary biliary cholangitis (PBC), and autoimmune pancreatitis (AIP).
ATF-2: activating transcription factor 2
bFGF: basic fibroblast growth factor
BrdU: 5’-bromo-2’-deoxyuridine
CD: Crohn’s disease
CDX2: caudal-type homeobox protein 2
DSS: dextran sulfate sodium
Dx: dexamethasone
E. coli: Escherichia coli
EGF: epidermal growth factor
EXT: exostoses
GATA6: GATA-binding factor 6
HGF: hepatocyte growth factor
HLTF: helicase-like transcription factor
IBD: inflammatory bowel diseases
KO: knockout
miR: microRNA
MZF1: myeloid zinc finger 1
NOD: nonobese diabetic
NON: non-obese non-diabetic
PAPs: pancreatitis-associated proteins
PARP: poly(ADP-ribose) polymerase
PI(3)K: 3-kinase
RAGE: receptor for advanced glycation endproducts
Reg: regenerating gene
RS: REG-related sequence
RTEF1: related transcriptional enhancer factor-1
siRNA: small interfering RNA
SOCS3: suppressor of cytokine signaling 3
SPF: specific pathogen-free
STAT3: signal transducer and activator of transcription 3
TEAD4: TEA domain transcription factor 4
Tg: transgenic
TNF: tumor necrosis factor
UC: ulcerative colitis
We are indebted to Hiroshi Okamoto and Michiaki Unno (Tohoku University), Marta Vives-Pi (Universitat Autònoma de Barcelona), Chikatsugu Tsuchida, Sumiyo Sakuramoto-Tsuchida, Asako Itaya-Hironaka, Shuhei Nakamura, Chiho Ohbayashi, and Akihiko Yoshizawa (Nara Medical University) for encouragement.
ST: Conceptualization, Writing—original draft, Writing—review & editing, Supervision, Validation. TU and MK: Writing—review & editing, Supervision, Validation. All authors read and approved the submitted version.
The authors declare that they have no conflict of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
This study is supported by in part by Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan grant number [08102003] and Japan Society for the Promotion of Science, and Japan Science and Technology Agency. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
© The Author(s) 2025.
Open Exploration maintains a neutral stance on jurisdictional claims in published institutional affiliations and maps. All opinions expressed in this article are the personal views of the author(s) and do not represent the stance of the editorial team or the publisher.
Copyright: © The Author(s) 2025. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Olha Denefil ... Khrystyna Loza
Weinan Du ... Juxiu Li
Alejandra Vargas ... David A. Johnson
Zeneng Wang ... Robert Koeth
