Affiliation:
1The Scientific Department of Oncoimmunology, NMRC of Oncology named after N.N. Petrov of MoH of Russia, 197758 St. Petersburg, Russia
ORCID: https://orcid.org/0000-0002-7826-4861
Affiliation:
1The Scientific Department of Oncoimmunology, NMRC of Oncology named after N.N. Petrov of MoH of Russia, 197758 St. Petersburg, Russia
2Department of Oncology, Pediatric Oncology and Radiation Therapy, St. Petersburg State Pediatric Medical University, 194100 St. Petersburg, Russia
Email: anovik@list.ru
ORCID: https://orcid.org/0000-0002-2430-4709
Affiliation:
1The Scientific Department of Oncoimmunology, NMRC of Oncology named after N.N. Petrov of MoH of Russia, 197758 St. Petersburg, Russia
3Department of Diagnostics of Functional Systems, St. Petersburg University, 199034 St. Petersburg, Russia
ORCID: https://orcid.org/0000-0003-4679-7281
Affiliation:
1The Scientific Department of Oncoimmunology, NMRC of Oncology named after N.N. Petrov of MoH of Russia, 197758 St. Petersburg, Russia
ORCID: https://orcid.org/0000-0003-4796-0386
Affiliation:
1The Scientific Department of Oncoimmunology, NMRC of Oncology named after N.N. Petrov of MoH of Russia, 197758 St. Petersburg, Russia
2Department of Oncology, Pediatric Oncology and Radiation Therapy, St. Petersburg State Pediatric Medical University, 194100 St. Petersburg, Russia
ORCID: https://orcid.org/0009-0002-1552-7684
Affiliation:
1The Scientific Department of Oncoimmunology, NMRC of Oncology named after N.N. Petrov of MoH of Russia, 197758 St. Petersburg, Russia
ORCID: https://orcid.org/0009-0001-9567-0802
Affiliation:
1The Scientific Department of Oncoimmunology, NMRC of Oncology named after N.N. Petrov of MoH of Russia, 197758 St. Petersburg, Russia
ORCID: https://orcid.org/0009-0000-6098-8661
Affiliation:
1The Scientific Department of Oncoimmunology, NMRC of Oncology named after N.N. Petrov of MoH of Russia, 197758 St. Petersburg, Russia
ORCID: https://orcid.org/0000-0002-3533-2721
Affiliation:
3Department of Diagnostics of Functional Systems, St. Petersburg University, 199034 St. Petersburg, Russia
ORCID: https://orcid.org/0000-0001-7641-956X
Affiliation:
1The Scientific Department of Oncoimmunology, NMRC of Oncology named after N.N. Petrov of MoH of Russia, 197758 St. Petersburg, Russia
ORCID: https://orcid.org/0000-0002-7472-4613
Explor Med. 2025;6:1001287 DOI: https://doi.org/10.37349/emed.2025.1001287
Received: December 01, 2024 Accepted: January 27, 2025 Published: February 27, 2025
Academic Editor: Apostolos Zaravinos, European University Cyprus, Cyprus
The article belongs to the special issue Molecular Diagnostics in Oncology
Aim: To assess the predictive and prognostic role of HLA class I expression in patients with melanoma (Mel), and soft tissue sarcomas (STS) treated with autologous dendritic cell vaccine (DCV) (CaTeVac).
Methods: From 2009 to 2023, 277 patients with Mel (143), and STS (134), received DCV at the N.N. Petrov National Medical Research Center of Oncology in adjuvant (78.3% and 14.9%) and therapeutic (21.7% and 85.1%) setting. HLA-typing was performed using a polymerase chain reaction with sequence-specific primers (PCR-SSP). Progression-free survival (PFS) and overall survival (OS) grouped by the presence of HLA alleles or HLA association rules were assessed using the Kaplan-Meier method (medians of survival in the month are presented).
Results: Higher OS (41.1 vs. 22.1, P = 0.026) and PFS (6.0 vs. 3.9, P = 0.045) were found in HLA-A heterozygous STS patients, while HLA-B homozygous patients showed better OS (36.4 vs. 87.2, P = 0.023). HLA-A heterozygous Mel patients showed lower PFS (8.3 vs. not reached, P = 0.013). Association rules analysis on HLA expression revealed 20 rules with high confidence, seven of which were associated with the survival. HLA-B*07 and HLA-C*07 (21.2 vs. 52.2), HLA-B*40 and HLA-C*03 (17.6 vs. 45.4), HLA-A*02 and HLA-B*07 and HLA-C*07 (16.8 vs. 47.0), HLA-A*02 and HLA-С*07 (17.6 vs. 41.1), HLA-B*40 and HLA-A*02 and HLA-C*03 (8.3 vs. 50.2) decreased OS in STS (P < 0.05). HLA-A*02 and HLA-B*07 and HLA-C*07 (3.2 vs. 6.0), HLA-B*40 and HLA-A*02 and HLA-C*03 (3.2 vs. 5.9) decreased PFS in STS patients (P < 0.05). HLA-B*35 and HLA-C*04 increased median OS in STS from 33.4 to 153.3 months.
Conclusions: HLA class I phenotype has a different impact on the survival in Mel and STS patients. The association rules based on HLA coexpression may have prognostic and predictive value. Further investigations of these parameters are warranted (The Trial Registration Number: NCT05539677).
Immunotherapy with the checkpoint inhibitors performed a revolution in the treatment of most solid tumors recently. This is particularly evident in cutaneous melanoma (Mel), which is known to be an immunologically dependent tumor. However, there has been little progress in other tumors, such as soft tissue sarcomas (STS). Other immunotherapy approaches remain promising and are still in the experimental stage, while evolving towards new technological levels.
Currently, there is a growing body of evidence on the clinical application of dendritic cell vaccines (DCVs) in various types of malignant tumors, including Mel, glioma, sarcoma, ovarian, bladder, kidney, pancreatic, hepatocellular, prostate, breast, non-small cell lung, colorectal, acute myeloid leukemia, lymphoma, and others [1–3]. The mechanism of action of all these products is the generation of a specific immune response against tumor antigens presented by dendritic cells (DCs), leading to the elimination of tumors by the immune system. While DCVs are being studied in phase I and II clinical trials for most tumors, phase III trials of its use in prostate cancer have been completed. Several phase III trials are ongoing in glioblastoma, uveal Mel, Mel, and kidney cancer [1, 3]. A meta-analysis has shown the overall survival (OS) gain in glioblastoma patients receiving DCV [4]. Another meta-analysis confirmed increased overall and progression-free survival (PFS) of patients with hepatocellular carcinoma on DCV therapy [5]. At the same time, a meta-analysis of six non-randomized clinical trials of DCV for prostate cancer failed to confirm the benefit of this type of therapy [6]. However, most studies have shown the high safety of DCV in cancer patients [5, 6]. The immune response after the administration of DCV has been also demonstrated [5, 6].
The first DCV was approved by the US. Food and Drug Administration (FDA) is Sipuleucel-T (Provenge®). This vaccine is an autologous mononuclear cell line of antigen-presenting cells (including DCs) loaded with the prostatic alkaline phospatase antigen, which has a high expression level on the surface of prostate cancer cells [7]. Phase I, II, and III clinical trials were conducted to assess the safety and efficacy of Sipuleucel-T in prostate cancer. These trials showed that the vaccine increased OS by 4.1 months and improved 3-year survival by 8.7% among patients with castration-resistant metastatic prostate cancer [8]. Other DCVs that are currently approved for the clinical practice include APCEDEN®, the Indian drug for the treatment of prostate, ovarian, non-small cell lung, and colorectal cancers, and CreaVax-RCC, a Korean vaccine for metastatic renal cell carcinoma, manufactured by JW CreaGene [9]. DCVax®-L, a product developed by Northwest Biotherapy in the United States, is currently undergoing a phase III clinical trial for patients with glioblastoma after primary surgery [10]. In this trial, patients receive DCV in combination with radiotherapy and chemotherapy.
We have developed an autologous DCV, called CaTeVac, which has several advantages over other DCVs. One of the main advantages is the use of highly immunogenic cancer-testis antigens (CTA) to activate DCs. These antigens are ideal for targeting immune responses in tumors, as they are strictly tumor-specific and do not cause autoimmune reactions since they are expressed in tumors of different histologies and are strictly tumor-specific. Several families of these antigens are known today, including MAGE, BAGE, GAGE, LAGE, HAGE, PASD1, SCP1, SEMG, SLLP, SPANXA, SSX1, PRAME and NY-ESO-1. The use of a wide variety of the above-mentioned CTA allows CaTeVac to target a broader range of tumor types than other dendritic cell solutions on the market. These other solutions typically use peptides or immune adjuvants to activate the immune system, limiting their effectiveness against certain tumors.
Today, it is clear that immunotherapy is effective for some patients. Researchers are working hard to find tools to predict which patients will benefit from this type of treatment. Many of these tools focus on human leukocyte antigens (HLA) expression. However, due to the complexity of the immune system, most of these efforts have not been successful in showing clinical benefits. Nevertheless, the important role of the HLA system in the body’s immune response to cancer requires further research to understand how to use this information to guide therapy. We conducted our study to assess the predictive and prognostic value of HLA class I expression in Mel and STS patients treated with CaTeVac. Due to the complexity of the HLA system and the large number of variables involved, we focused on assessing the heterogeneity of HLA class I alleles and patterns in HLA coexpression.
From 2009 to 2023, a total of 277 patients with Mel (143) or STS (134) received CateVac at the N.N. Petrov National Medical Research Center of Oncology in several clinical trials that were aggregated in the REGATA registry (NCT05539677) [11].
Written informed consent was obtained from all participants in the study. The main inclusion criteria for the study were: age over 18 years, histologically confirmed diagnosis of either STS or Mel, stage II–IV disease, eastern cooperative oncology group (ECOG) status 0–2, conduction of previous standard therapy of the disease, the satisfactory function of internal organs and bone marrow, absence of common contraindications to systemic drug treatment.
Exclusion criteria included pregnancy or breastfeeding, decompensation of concomitant chronic diseases, therapy with systemic corticosteroids and/or other immunosuppressive drugs within 4 weeks before therapy starts, high likelihood of their use during the study for the treatment of intercurrent pathology, acute infectious process, inflammation or scarring of the skin at the sites of the proposed vaccine administration, autoimmune diseases (excluding vitiligo), and psychiatric diseases; study discontinuation criteria include radiologically confirmed tumor progression, withdrawal of informed consent, patient’s failure to follow the study procedures, and development of diseases or conditions that prevent the patient from continuing to participate in the study. Therapy reinduction was permitted as a separate line of therapy. Patients were registered as separate subjects for survival analysis in this case. Only unique patients were included in the HLA typing analysis. The characteristics of the patients included in the study are presented in Table 1.
Patient’s characteristics
| Parameter | Mel | STS | |||
|---|---|---|---|---|---|
| Adj | Met | Adj | Met | ||
| N | 112 | 31 | 20 | 114 | |
| Age, years | Median | 52 | 52 | 38.5 | 47 |
| Range | 18–85 | 18–84 | 25–65 | 18–79 | |
| Sex | Male | 50 | 12 | 9 | 37 |
| Female | 62 | 19 | 11 | 77 | |
| TNM stage at inclusion | II | 16 | 0 | 6 | 17 |
| III | 57 | 2 | 3 | 20 | |
| IV | 39 | 29 | 11 | 77 | |
| Median follow-up, months | 42 | 9.9 | 51 | 11.1 | |
| Time from the last patient’s first dose, months | 60 | 37 | 50 | 29 | |
Mel: melanoma; STS: soft tissue sarcomas; Adj: adjuvant setting; Met: metastatic setting; N: number; TNM: tumor, nodus and metastasis
The patient received CaTeVac as described previously [12] in the adjuvant setting (Adj) or metastatic setting (Met).
CateVac was produced by differentiation of DCs from the adherent monocyte fraction (CD14+) of the peripheral blood of patients with malignant neoplasms under conditions of 37°C, 5% CO2, and 98% humidity in adherent culture vials (Sarstedt, Germany), using a balanced serum-free medium “Cell-Gro DC” (CellGenix, Germany). Growth and differentiation factors (produced by CellGenix, Germany), including GM-CSF (72 ng/mL) and IL-4 (20 ng/mL), were used and applied on days 1, 3, and 5 of cultivation.
For the loading and specific activation of immature DC (CD14– CD1a+), a cocktail of allogeneic tumor cell lines lysates, “IRTAN”, was used. This is a cell product for the load of DCs that consists of the broad spectrum of CTA-expressing cells lysate and lacks specific immunosuppressive action on the immune cells [13]. The genetic authenticity of the product was confirmed by STR-analysis, LLC “GORDIZ”, Moscow. Cells used for IRTAN had high expression of GAGE-1, HAGE, NY-ESO-1, MAGEA1, PASD1, SCP1, SEMG1, SLLP1, SPANXA1, SSX1, and PRAME. Lysed cells were added to immature DC in a 3:1 ratio [13]. After 48 h, DC (CD1a– CD83+) were collected, and precipitated by centrifugation. We studied the expression of lineage-specific and differentiation antigens on both immature and mature DC using antibodies to DC surface antigens directly labeled with fluorochromes (anti-CD83-PE-Cy7, anti-CD1a-APC, anti-CD80-APC-Cy7, anti-CD86-PerCP-Cy5.5, anti-CD40-PE, anti-CD14-FITC, anti-CCR7-BV421, anti-HLA-DR-APC-Cy7, anti-CD209-PerCP-Cy5.5) using BD FACS Canto II laser flow cytofluorimetry (BD, USA) to confirm product quality.
CaTeVac was injected intradermally paravertebrally at a dose of 9–15 million cells per injection with an interval of 2–4 weeks. All patients received CaTeVac irrespectively to cancer-testis antigen expression since different families of these antigens are expressed in all tumors [14]. CaTeVac was injected three days after intravenous injection of 300 mg cyclophosphamide for immune modulation. DC was injected on days 1, 14, 35, and 56, and then monthly.
In all patients, a radiology assessment was performed every eight weeks according to RECIST 1.1 criteria (only the presence of disease progression was evaluated for the adjuvant group) [15]. Patients received treatment until disease progression, grade 3–4 toxicity, or one year of treatment in the Adj, whichever came first.
PFS was measured from the start of therapy (surgery in the Adj or CaTeVac in the Met) until progression by RECIST 1.1 criteria [16]. OS was measured from the same timepoint as PFS until death from any cause.
Peripheral blood mononuclear cells from the included patients were used for HLA class I genotyping using the polymerase chain reaction with sequence-specific primers (PCR-SSP) method. DNA was isolated from the mononuclear cells using DNA isolation kits (PROTRANCE DNA Box 500, Protrans, Germany) followed by DNA concentration testing using a Quantus fluorimeter (Promega, USA). HLA typing was performed using Protrans reagents (Germany) for the HLA-A/B/C loci, according to the manufacturer’s instructions. The results of the HLA typing are presented in Supplementary material.
Statistical analysis was performed with R v4.4.2 programming language [17]. Survival analysis was performed using “survival” [18] and “survminer” [19] packages. We used logrank test to compare survival distributions. Association rules analysis was performed using “arules” and “arulesViz” packages [20].
The prognostic and predictive value of HLA molecule expression has different prognostic significance for patients with Mel and STS. HLA-A heterozygosity has an opposite impact on PFS (Figure 1) in these two groups. In patients with STS, those who were heterozygous by HLA had a median PFS of 6.0 monthes, compared to 3.9 months for the rest of the patients (P = 0.045). For Mel patients, the median PFS was 8.63 months for heterozygous patients and not reached for homozygous patients (P = 0.013). OS analysis (Figure 1) confirmed these findings in patients with STS. The median OS was 41.1 months for heterozygous patients, compared to 22.1 months for the others (P = 0.026). No significant differences were found for OS in Mel patients based on HLA-A heterozygosity (P = 0.579).
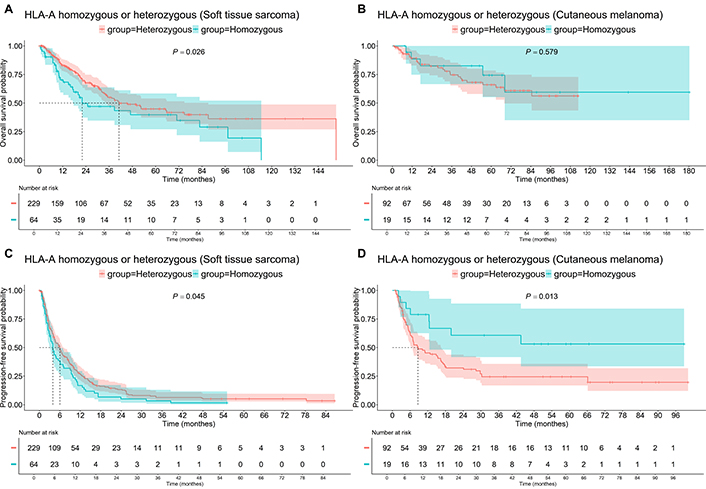
Survival of patients with homozygous or heterozygous “HLA-A” status. A: Soft tissue sarcoma, OS; B: cutaneous melanoma, OS; C: soft tissue sarcoma, PFS; D: cutaneous melanoma, PFS. OS: overall survival; PFS: progression-free survival
Homozygosity at the HLA-B locus was associated with improved OS in patients with STS (Figure 2). The median OS was 36.4 months for heterozygous patients compared to 87.2 months for the rest of the group (P = 0.023). However, there were no significant differences in OS among Mel patients (P = 0.166). Additionally, no impact of HLA-B heterozygosity on PFS was observed in either Mel (P = 0.687) or STS (P = 0.728) patients (Figure 2).
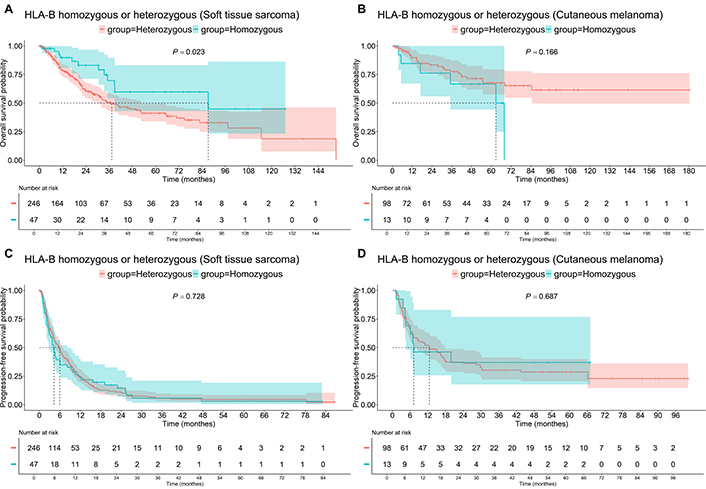
Survival of patients with homozygous or heterozygous “HLA-B” status. A: Soft tissue sarcoma, OS; B: cutaneous melanoma, OS; C: soft tissue sarcoma, PFS; D: cutaneous melanoma, PFS. OS: overall survival; PFS: progression-free survival
Thus, we have demonstrated the impact of HLA class I heterozygosity on the clinical outcome of DCV patients.
To find HLA-I (A, B, and C) combinations in patients with melanoma and STS who received DCV. These combinations have prognostic and predictive value. An association rule can be expressed in the form of X => Y, meaning that if an observation has item X, there is a high likelihood that item Y will also be present (more formally, this is an estimate of the conditional probability of Y given X).
It should be noted that X can be a set of items. For example, HLA-A*01 and HLA-B*08 imply HLA-C*07. There are three main measures related to association rule analysis: support, confidence, and lift. Support measures how frequently an item or a set of items appears in the data. Confidence is the ratio of the support of X and Y to the support of X alone, which is an estimate of the conditional probability of Y given X. Lift is used to compare the observed confidence of X and Y with their expected confidence. In terms of support, this is the support of X and Y divided by the support of both X and Y. We used a confidence threshold of 70% to find the association rules.
Twenty association rules were found, and they are summarized in Figure 3. The detailed characteristics of each rule can be found in Table 2.
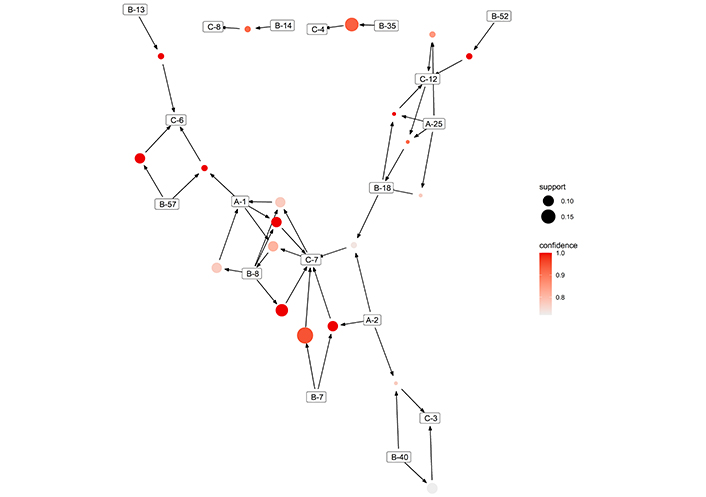
Graph of found associative rules between HLA with confidence higher than 70% via A-Priori algorithm in all included patients
Association rules statistics of HLA-I (A, B, and C) combinations of Mel and STS patients who received DCV immunotherapy
| Lhs1 | Rhs2 | Support | Confidence | Cov3 | Lift | Count4 | |
|---|---|---|---|---|---|---|---|
| 1 | HLA-B*13 | HLA-C*06 | 0.0598 | 1.0000 | 0.0598 | 4.000 | 1 |
| 2 | HLA-B*52 | HLA-C*12 | 0.0598 | 1.0000 | 0.0598 | 4.182 | 11 |
| 3 | HLA-A*25 | HLA-B*18 | 0.0543 | 0.7692 | 0.0707 | 4.423 | 10 |
| 4 | HLA-A*25 | HLA-C*12 | 0.0597 | 0.8462 | 0.0707 | 3.538 | 11 |
| 5 | HLA-B*14 | HLA-C*08 | 0.0598 | 0.9167 | 0.0652 | 10.542 | 11 |
| 6 | HLA-B*57 | HLA-C*06 | 0.0924 | 1.0000 | 0.0923 | 4.000 | 17 |
| 7 | HLA-B*08 | HLA-A*01 | 0.0924 | 0.7727 | 0.1196 | 3.3852 | 17 |
| 8 | HLA-B*08 | HLA-C*07 | 0.1196 | 1.0000 | 0.1196 | 2.0909 | 22 |
| 9 | HLA-B*40 | HLA-C*03 | 0.0978 | 0.7200 | 0.1358 | 3.5805 | 18 |
| 10 | HLA-B*35 | HLA-C*04 | 0.1413 | 0.9286 | 0.1522 | 4.4962 | 26 |
| 11 | HLA-B*07 | HLA-C*07 | 0.1902 | 0.9459 | 0.2010 | 1.9779 | 35 |
| 12 | HLA-B*18, HLA-A*25 | HLA-C*12 | 0.0543 | 1.0000 | 0.0543 | 4.1818 | 10 |
| 13 | HLA-C*12, HLA-A*25 | HLA-B*18 | 0.0543 | 0.9091 | 0.0598 | 5.2273 | 10 |
| 14 | HLA-B*57, HLA-A*01 | HLA-C*06 | 0.0597 | 1.0000 | 0.0598 | 4.0000 | 11 |
| 15 | HLA-B*08, HLA-A*01 | HLA-C*07 | 0.0923 | 1.0000 | 0.0924 | 2.0909 | 17 |
| 16 | HLA-B*08, HLA-C*07 | HLA-A*01 | 0.0924 | 0.7727 | 0.1196 | 3.3853 | 17 |
| 17 | HLA-C*07, HLA-A*01 | HLA-B*08 | 0.0924 | 0.8095 | 0.1141 | 6.7706 | 17 |
| 18 | HLA-B*40, HLA-A*02 | HLA-C*03 | 0.0543 | 0.7692 | 0.0707 | 3.8254 | 10 |
| 19 | HLA-B*18, HLA-A*02 | HLA-C*07 | 0.0598 | 0.7333 | 0.0815 | 1.5333 | 11 |
| 20 | HLA-B*07, HLA-A*02 | HLA-C*07 | 0.0924 | 1.0000 | 0.0924 | 2.0909 | 17 |
1 Lhs is left-hand-side or X in the association rule; 2 Rhs is right-hand-side or Y in the association rule; 3 Coverage is a proportion of observations in the full dataset that comply with the rule; 4 Count of the observations with the rule. Mel: melanoma; STS: soft tissue sarcomas; DCV: dendritic cell vaccine
We compared the survival of patients who had all the HLA loci present in each association to the rest of the patients. The results of these comparisons are presented in Figures 4–9.
The median OS for patients with HLA-В*07 and С*07 was 21.2 months compared to 52.5 months for the rest of the patients, P < 0.001 (Figure 4). However, the same trend did not reach a prespecified level of significance in the Mel group (P = 0.091), probably due to the smaller number of patients in this group. No significant association with PFS was found for this rule in either group (P > 0.05).
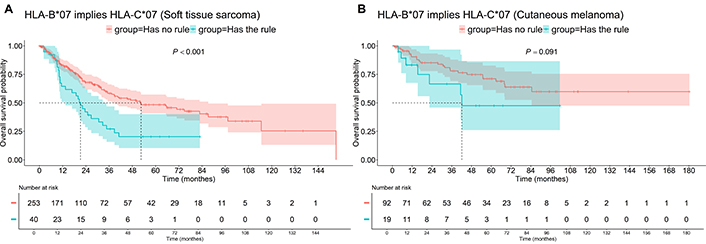
OS of patients with “HLA-B*07 implies HLA-C*07” association rule, i.e., having HLA-B*07 and HLA-C*07. A: Soft tissue sarcoma; B: cutaneous melanoma. OS: overall survival
The presence of HLA-В*35 and С*04 in STS patients increased median OS from 33.4 to 153.3 months (P < 0.001), while having no impact on OS in Mel patients (Figure 5). Neither impact was found in the PFS analysis.
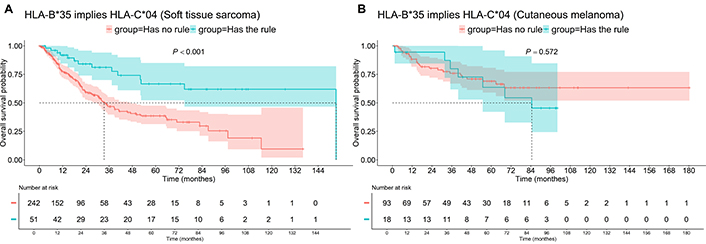
OS of patients with “HLA-B*35 implies HLA-C*04” association rule, i.e., having HLA-B*35 and HLA-C*04. A: Soft tissue sarcoma; B: cutaneous melanoma. OS: overall survival
For patients with HLA-В*40 and HLA-С*03 (Figure 6), the OS median for STS patients was 17.6 months, compared to 45.4 months for the rest of the patients (P = 0.002). A similar trend was observed in the PFS of STS patients (Figure 6), with a median decrease from 5.7 to 3.9 months (P = 0.006). For Mel patients, the association of HLA-В*40 and HLA-С*03 did not show any significant impact on survival.
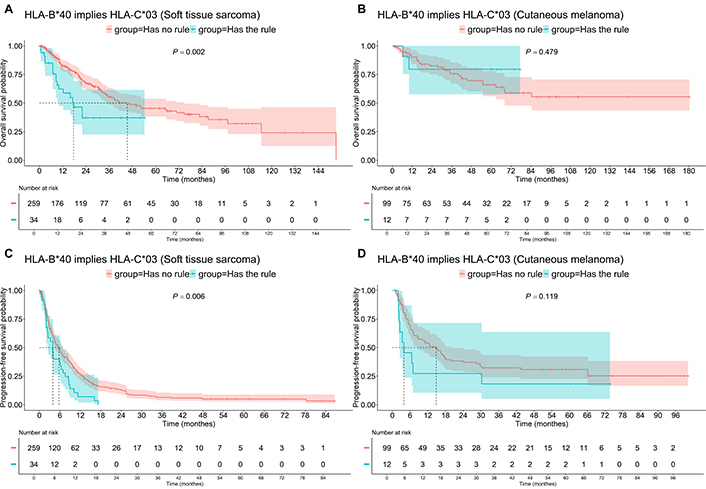
Survival of patients having “HLA-B*40 implies HLA-C*03” association rule, i.e., having HLA-B*40 and HLA-C*03. A: Soft tissue sarcoma, OS; B: cutaneous melanoma, OS; C: soft tissue sarcoma, PFS; D: cutaneous melanoma, PFS. OS: overall survival; PFS: progression-free survival
The association of HLA-A*02, HLA-B*07, and HLA-С*07 in STS patients was found to be worse, decreasing the median OS from 47 months to 16.8 months (P = 0.001, Figure 7A). A similar trend was observed when HLA-A*02 and HLA-С*07 were associated with HLA-B*18 in STS patients (Figure 8), where the median OS decreased from 41.1 months to 17.6 months (P = 0.028). However, the combination of HLA-A*02, HLA-B*18, and HLA-С*07, but not the combination of HLA-A*02, HLA-B*07, and HLA-С*07 decreased the median PFS in STS patients from 6.0 to 3.2 months (P = 0.015). No significant survival impact of any of these HLA allele combinations was observed in Mel patients.
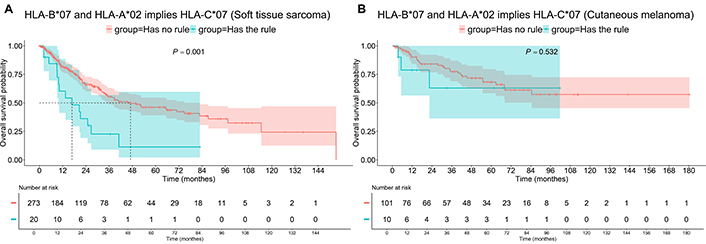
OS of patients having “HLA-B*07 and HLA-A*02 implies HLA-C*07” association rule, i.e., having HLA-B*07 and HLA-A*02 and HLA-C*07. A: Soft tissue sarcoma; B: cutaneous melanoma. OS: overall survival
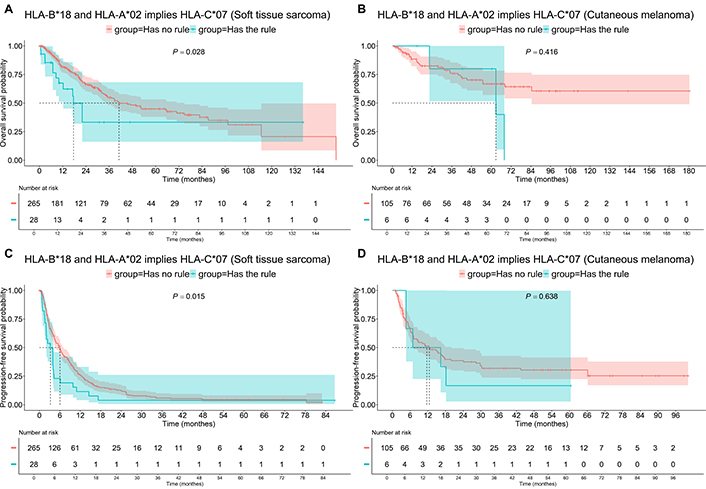
Survival of patients having “HLA-B*18 and HLA-A*02 implies HLA-C*07” association rule, i.e., having HLA-B*18 and HLA-A*02 and HLA-C*07. A: Soft tissue sarcoma, OS; B: cutaneous melanoma, OS; C: soft tissue sarcoma, PFS; D: cutaneous melanoma, PFS. OS: overall survival; PFS: progression-free survival
The association of HLA-A*02, HLA-B*40, and HLA-С*03 with worse outcomes was only observed in STS patients (Figure 9). The median OS for these patients was 8.6 months compared to 50.2 months (P < 0.001) for the rest of the patients. The median PFS was 3.2 months for these patients, compared to 5.9 months for the other patients (P = 0.002).
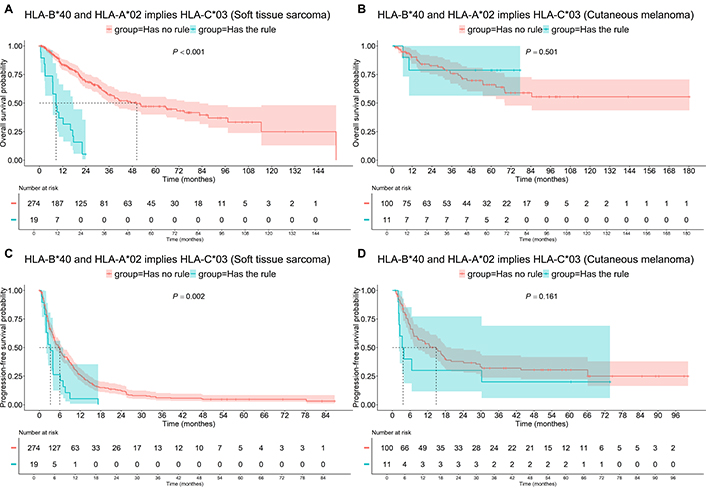
Survival of patients having “HLA-B*40 and HLA-A*02 implies HLA-C*03” association rule, i.e., having HLA-B*40 and HLA-A*02 and HLA-C*03. A: Soft tissue sarcoma, OS; B: cutaneous melanoma, OS; C: soft tissue sarcoma, PFS; D: cutaneous melanoma, PFS. OS: overall survival; PFS: progression-free survival
Our data provides evidence of different prognostic and predictive values of HLA phenotypes in Mel and STS. These differences can be both quantitative and qualitative. For the majority of studied parameters, a prognostic role rather than a predictive one can be assumed. Further studies are required to confirm these findings and apply them to patient selection in the DCV trials.
The state of the patient’s immune system and the ability of the therapy to impact the effector and suppressor components of immunity determine the prognosis of the disease and the success of various types of anti-tumor therapy, including targeted therapy, chemotherapy, and immunotherapy [21]. Reliable immunological predictors remain a significant unmet medical need now. Many tissues and peripheral blood-based immunological markers have been studied for this purpose, but no single definitive approach has been established.
The HLA plays a crucial role in the immune system, determining the presentation of antigens to T-cells and facilitating the recognition and destruction of foreign substances, including tumor cells. HLA is located on the short arm of chromosome 6, and consists of about 4 million nucleotide pairs in more than 200 genes. Many HLA genes are highly polymorphic, and only 40% of them encode proteins involved in the immune response [22]. HLA class I molecules bind and export peptides derived from tumor antigens to the surface of the antigen-presenting cells, where they are recognized by CD8+ T-lymphocytes. These lymphocytes are then activated and become capable of destroying tumor cells. This process depends on two main factors: the efficiency of binding of tumor epitopes to HLA class I molecules and the interaction between this complex and the T-cell receptor, leading to a cytotoxic response against the target cell. Polymorphism of HLA molecules leads to differences in the ability to bind peptides. As a result, certain combinations of HLA class I alleles may have limited interaction with tumor antigens, especially in the context of immune evasion strategies employed by tumors [22, 23].
Although there is relatively little known about the binding capacity and immunogenicity of specific HLA alleles to tumor antigens, it is assumed that alleles that have a high affinity for tumor-associated antigens could enhance the efficacy of immunotherapy with immune checkpoint blockade or antitumor vaccines, particularly those based on DCs. The varying ability of HLA proteins to effectively present peptides may specifically decrease or increase the antitumor immune response. This was confirmed by C.H. Lee et al. [24], who showed the role of HLA diversity was shown. Other efforts to generalize the function of HLA alleles through the classification of superalleles [25] have not been successful in predicting response to immunotherapy [26]. The frequency of co-expression of HLA class I alleles is mostly used for transplantation purposes [27]. We analyzed the occurrence of major loci of HLA class I loci(A/B/C) in patients with Mel and STS and found associative relationships between certain alleles of individual loci and their association with OS and PFS. Four of the six combinations of HLA class I alleles were associated with worse OS in STS patients. The HLA-A*02, B*18, and C*07 combination was associated with decreased OS and PFS in STS patients. The presence of the HLA-A*02, B*40, and C*03 combination was associated with worse OS of Mel and SMT patients as well as decreased PFS in STS patients. Certain HLA alleles have been shown to influence cancer patients’ survival outcomes, and therefore, their role as potential prognostic biomarkers is receiving increasing attention [28]. Moreover, a prognostic role for HLA alleles in patients receiving immune checkpoint inhibitors has been demonstrated [29]. However, there is currently a lack of data on the prognostic significance of the presence of specific HLA alleles, especially in patients treated with antitumor vaccines. The results of studies on sarcomas are particularly scarce. For example, Rosenbaum et al. [30] found that the HLA-A*02 haplotype was marginally associated with shorter OS in patients with synovial sarcoma patients who received immunotherapy with genetically modified NY-ESO-1-specific T cells as part of a clinical trial (HR 1.95, 95% CI 0.995–3.813, P = 0.052).
We found that HLA-B*07 and HLA-C*07 allele combination is associated with a worse OS in patients with STS. Similar results were obtained by Lotem et al. [31], who assessed the efficacy of an allogeneic Mel vaccine composed of three cell lines, each corresponding to at least one allele of the HLA-A and -B loci in the recipient. They found that patients with HLA-B*07 expression had a lower OS. Interestingly, HLA-B*07 was also identified as a prognostic factor for an increased risk of breast cancer [32].
Genetic instability can lead to somatic mutations, which must be controlled by the immune system. Malignant neoplasms are accompanied by disorders in antigen-presenting cells and T-lymphocyte interactions. T cell responses to peptides depend critically on binding and presentation by HLA class I and class II molecules. Certain HLA antigens and their combinations, called haplotypes, may be involved in “immune escape” processes. Tumor cells can evade immune surveillance due to the inability of specific HLA proteins to bind tumor-associated peptides adequately, resulting in reduced or absent presentation of these peptides and contributing to disease progression.
It was found that Mel patients who were homozygous for HLA-A had a better OS and increased PFS. On the other hand, STS patients who were heterozygous for HLA-A showed a more favorable outcome. Homozygosity for HLA-B was also associated with better survival in the STS group. However, homo- or heterozygosity at the HLA-C locus did not significantly affect survival in either group. It should be noted that previous studies have yielded conflicting results regarding the prognostic value of homo- and heterozygosity of HLA class I loci. For example, Abed A. et al. (2024) [33] found no association between HLA-I/-II homozygosity and clinical outcomes in a population of patients with non-small cell lung cancer treated with pembrolizumab and chemotherapy. However, the presence of HLA-A*01 was associated with unfavorable PFS [hazard ratio (HR) = 2.32; 95% confidence interval (CI) 1.13–4.77; P = 0.022] and worse OS (HR = 2.86, 95% CI 1.06–7.70; P = 0.038). The presence of HLA-B*27 was associated with improved PFS (HR = 0.35; 95% CI 0.18–0.71; P = 0.004) and a trend toward better OS [33]. In contrast, an earlier study by Chowell et al. [29] showed that maximal heterozygosity at HLA class I loci (A/B/C) improved the OS of Mel patients after treatment with immune checkpoint inhibitors, compared to patients who were homozygous for at least one HLA locus. Furthermore, patients with the HLA-B*44 super-type had longer survival, while the HLA-B*62 super-type or somatic loss of heterozygosity in the HLA class I region was associated with an unfavorable outcome.
Our study has several limitations that may have biased its results. For example, we did not take into account the specific therapy setting, stage of disease, or other potential prognostic factors. Additionally, our dataset was not large enough to perform a multifactorial analysis or to use other techniques to address potential imbalances. The primary aim of our study was to explore possible associations between HLA phenotype and the efficacy of CaTeVac therapy. However, all findings from this study should be confirmed in further, larger, and more robust trials.
Clearly, more in-depth research is needed on larger samples of patients receiving different types of immunotherapy. It is also essential to consider the biological characteristics of various malignancies, which may influence the specificity of immune interactions. Further investigations into the mechanisms of the HLA system in cancer are crucial to using HLA phenotypes as predictive and prognostic biomarkers in clinical practice.
Adj: adjuvant setting
CI: confidence interval
DCs: dendritic cells
DCV: dendritic cell vaccine
HLA: human leukocyte antigens
HR: hazard ratio
Mel: melanoma
Met: metastatic setting
OS: overall survival
PCR-SSP: polymerase chain reaction with sequence-specific primers
PFS: progression-free survival
STS: soft tissue sarcomas
СTA: cancer-testis antigens
The supplementary material for this article is available at: https://www.explorationpub.com/uploads/Article/file/1001287_sup_1.xlsx.
TLN: Supervision, Project administration, Validation. AVN: Conceptualization, Data curation, Software. DVG: Formal analysis, Methodology, Visualization. ABD: Writing—review & editing, Validation. PAS: Writing—original draft, Data curation. AVG and MAN: Resources, Investigation. NAE: Writing—review & editing, Data curation. AVO: Data curation, Formal analysis. IAB: Project administration, Validation. All authors read and approved the submitted version.
The authors declare that they have no conflicts of interest.
The study was approved by the ethics committee of the N.N. Petrov National Medical Research Center of Oncology Protocol No. 2 dated 20.02.2020.
Informed consent to participate in the study was obtained from all participants.
Not applicable.
The datasets for this manuscript are not publicly available, since informed consent was not obtained from the participants for data sharing with third parties. Requests for access to the aggregated data from the study datasets should be directed to the corresponding author.
Not applicable.
© The Author(s) 2025.
Open Exploration maintains a neutral stance on jurisdictional claims in published institutional affiliations and maps. All opinions expressed in this article are the personal views of the author(s) and do not represent the stance of the editorial team or the publisher.
Copyright: © The Author(s) 2025. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
View: 2230
Download: 38
Times Cited: 0
Viktoria Borobova ... Sergey Kovalenko
Noor Mey Wardhani ... Bulkis Natsir
Evgeny Imyanitov, Anna Sokolenko
