Affiliation:
Department of Pharmacy, Quaid-i-Azam University, Islamabad 45320, Pakistan
ORCID: https://orcid.org/0009-0009-9540-0747
Affiliation:
Department of Pharmacy, Quaid-i-Azam University, Islamabad 45320, Pakistan
ORCID: https://orcid.org/0009-0008-4847-1799
Affiliation:
Department of Pharmacy, Quaid-i-Azam University, Islamabad 45320, Pakistan
ORCID: https://orcid.org/0009-0002-1317-3819
Affiliation:
Department of Pharmacy, Quaid-i-Azam University, Islamabad 45320, Pakistan
ORCID: https://orcid.org/0009-0005-6860-1675
Affiliation:
Department of Pharmacy, Quaid-i-Azam University, Islamabad 45320, Pakistan
Email: sameenabbas@bs.qau.edu.pk
ORCID: https://orcid.org/0000-0002-7260-3973
Explor Neuroprot Ther. 2024;4:240–250 DOI: https://doi.org/10.37349/ent.2024.00080
Received: February 27, 2024 Accepted: April 18, 2024 Published: May 28, 2024
Academic Editor: Agnese Secondo, “Federico II” University of Naples, Italy
Epilepsy, a complex neurological disorder, is influenced by intricate interactions within cortical, hippocampal, or thalamocortical neuronal networks, presenting a genetically complex condition with non-Mendelian inheritance patterns. This complexity is underscored by the involvement of numerous “susceptibilities” or “modifier” genes, complicating the assessment of risk and therapy outcomes. A critical inquiry in epilepsy treatment involves understanding how genetic diversity impacts treatment strategies and efficacy. Pharmacogenomic advancements have elaborated the connection between genetic variants and antiseizure medication (ASM) safety and response, marking a shift towards precision medicine in epilepsy care. Notably, genetic screening for variants such as HLA-B*1502 and HLA-A*3101 has demonstrated significant efficacy in preventing severe hypersensitivity reactions, including toxic epidermal necrolysis (TEN) and Stevens-Johnson syndrome (SJS), particularly among specific ethnic populations. However, putting pharmacogenomic discoveries into clinical practice faces numerous challenges, including educational, legal, and economic barriers, emphasizing the need for broader acceptance and integration of pharmacogenomic data. This review synthesizes recent studies on pharmacogenomics in epilepsy, highlighting the current advances and prospects for personalizing epilepsy treatment through genetic insights, aiming to enhance ASM safety, reduce adverse effects, and improve treatment outcomes. Through a comprehensive examination of the genetic basis of epilepsy and its influence on pharmacotherapy, this review endeavors to contribute to the evolving landscape of precision medicine in epilepsy care, advocating for a more individualized and effective treatment approach.
Epilepsy affects over 0.5% of people worldwide. It comes with a range of challenges, including different types of seizures and how differently patients respond to antiseizure medications (ASMs) [1]. This variation makes it difficult to find the right treatment for everyone, often leading to uncontrolled seizures and side effects. This is why personalized treatments are so crucial. Pharmacogenomics, a key area in precision medicine, can customize ASM therapy by considering each person’s genetic makeup. This approach aims to improve treatment results, marking a new era in epilepsy care [1, 2]. Recent progress has highlighted how genetics play a role in determining the safety and efficacy of ASMs. Certain studies have identified specific genetic variations that affect how ASMs work and how the body processes them. These findings include well-known pathways, like the mTOR signalling pathway and SCN1A gene. The impact of T-type calcium channels and their connection to CACNA1H mutations, along with their various manifestations, has contributed to advancements in understanding the diverse effects of ASMs on individuals [3].
Drug-resistant epilepsy (DRE) is a major challenge that affects approximately one-third of people with epilepsy. It occurs when seizures continue even after trying two or more ASMs. Genetics, specifically variations in genes related to drug transport and ion channels, plays a significant role in DRE. It is crucial to understand these genetic factors to develop more effective and personalized treatments for this condition [4, 5].
Despite significant advancements in our understanding of epilepsy, there are still gaps in our knowledge regarding its genetic aspects. These gaps can impede personalized care and create barriers to treatment advancements. This article aims to address these gaps by providing a comprehensive review of the latest findings in epilepsy pharmacogenomics. It delves into how genetic factors influence the response to ASM and highlights the clinical implications of these findings. By exploring these insights, pharmacogenomics shows great potential in transforming epilepsy treatment by making drugs more personalized, reducing side effects, and enhancing treatment effectiveness.
Epilepsy can manifest in various ways and may not respond to ASMs in the same way for everyone. This highlights the importance of personalized treatment strategies. Pharmacogenomics makes it possible to customize ASM therapy based on an individual’s genetic profile, leading to better treatment outcomes and signaling a new era in epilepsy care [6]. This is particularly important for patients with DRE. Studies have shown the crucial role of genetic factors in determining ASM effectiveness and safety, making pharmacogenomics a key player in customizing epilepsy management [4].
The literature on epilepsy’s pathophysiology shows that it is a complex condition with a strong genetic component [7]. Therefore, epilepsy care should adopt a precision medicine approach that takes into account an individual’s unique genetic makeup. Genome-wide association studies have identified genetic markers that influence how patients respond to ASMs in genetically generalized epilepsies [3]. This highlights the potential of pharmacogenomics to revolutionize epilepsy treatment by enabling personalized drug therapy.
Understanding the difference between polymorphisms and disease-associated mutations is essential to comprehend genetic influences on drug response and disease susceptibility. Polymorphisms, which are prevalent in over 1% of the population, are single or multiple nucleotide variations that significantly affect drug action due to their genetic substrates. In contrast to these prevalent variations, mutations are infrequent but can produce a diverse range of phenotypic manifestations, affecting protein function and subsequently influencing drug interactions. This complexity highlights both the challenges and potential of genetically driven pharmacology for developing tailored treatments [8].
DRE challenges a significant fraction of epilepsy patients, with about one-third not responding to multiple ASMs. The exploration of genetic variations, including those in the ABCB1 and CACNA1H genes, reveals the complex genetic basis of DRE and emphasizes the need for individualized approaches [9]. As research continues to uncover the genetic factors associated with the response to ASMs, the integration of pharmacogenomic data into clinical practice is becoming increasingly important. This presents opportunities for pharmacogenomics to guide the development of targeted therapies for epilepsy, especially in the context of DRE [10].
Pharmacogenomics has made significant progress in promoting neuroprotection for epilepsy patients. The goal is to not only improve seizure management but also preserve neurological function. Metformin is a promising example of a drug that has demonstrated neuroprotective effects in epilepsy. By activating AMP-activated protein kinase (AMPK) and possessing antioxidant properties, it has the potential to be an effective treatment beyond traditional ASMs [11]. Moreover, targeting neuroinflammatory pathways in epileptogenic brain areas suggests a new direction in overcoming the limitations of current medications [12]. Novel therapeutic strategies like rhein, known for its anti-inflammatory properties, demonstrate anticonvulsant and neuroprotective effects by inhibiting specific signaling pathways and reducing cytokine secretion [13]. These developments underline the revolutionary potential of pharmacogenomics in evolving epilepsy treatment into a more individualized and neuroprotective approach.
Recent advancements in pharmacogenomics have significantly improved our understanding of how genetic variations influence the pharmacokinetics and pharmacodynamics of ASMs, thereby affecting individual responses and resistance to therapy as shown in Figure 1.
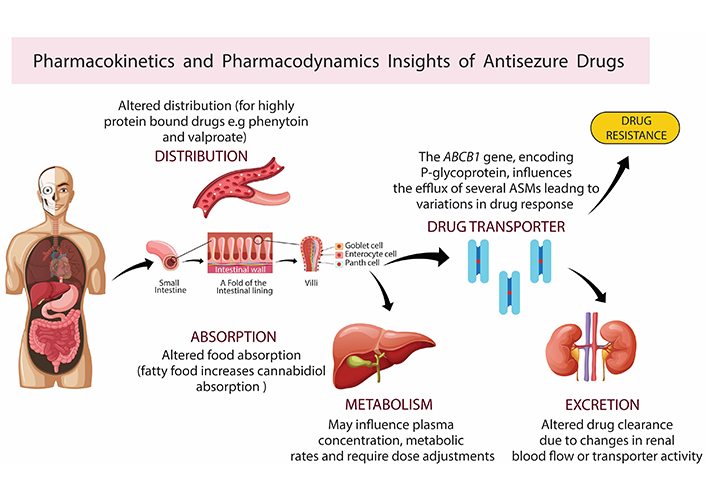
Pharmacokinetics and pharmacodynamics insights of antiseizure medications (ASMs). Pharmacokinetic and dynamic parameters, influenced by body condition and genetic variants like cytochrome P450 enzymes, P-glycoprotein, and others, impact drug absorption, distribution, metabolism, and excretion. Variants in target receptors such as voltage-gated sodium channels (e.g., SCN1A, SCN2A) alter drug efficacy, potency, and adverse effects, and may induce drug resistance. Created with BioRender.com
The metabolism of ASMs is profoundly affected by genetic polymorphisms in the CYP2C9 and CYP2C19 genes, with significant implications for drug levels, efficacy, and adverse effects [14]. Recent studies have highlighted the influence of CYP2C93 and CYP2A64 variants on plasma concentrations of valproic acids, suggesting that carriers of these alleles might require dosage adjustments to maintain therapeutic efficacy and avoid toxicity as shown in Table 1.
Genetic determinants of ASMS pharmacokinetics
| Antiepileptic drug | Genetic polymorphisms | Implications | Reference(s) |
|---|---|---|---|
| Valproic acid | CYP2C93, CYP2A64 | Influence on plasma concentrations may require dosage adjustments | [14] |
| Phenytoin | CYP2C92, CYP2C93, CYP2C19*2 | Reduced metabolism rate, unclear effects on CYP2C19*2, need for dosage adjustments.CYP2C19 mutations: Carriers may require reduced maintenance doses.CYP2C93: Associated with increased risk of severe cutaneous adverse reactions | [15–17] |
Research conducted on the metabolism of phenytoin among Middle Eastern and North African populations found that the CYP2C9*2 and CYP2C9*3 variants significantly reduced the rate of metabolism. However, the effects of the CYP2C19*2 and CYP2C9*3 variants on metabolism were not clear, indicating that the function of CYP2C9 variants plays a crucial role in determining the pharmacokinetics of phenytoin [15, 16]. Furthermore, recent studies have emphasized the importance of genetic testing in optimizing phenytoin therapy. It has been revealed that individuals who are homozygous or heterozygous carriers of CYP2C19 mutations require significantly reduced maintenance doses. In addition, a meta-analysis focused on the relationship between the CYP2C93 variant and the risk of phenytoin-induced Stevens-Johnson syndrome (SJS) and toxic epidermal necrolysis (TEN) has highlighted the clinical significance of CYP2C9 genotyping in minimizing the risk of severe cutaneous adverse reactions. Table 1 depicts the overall summary of genetic determinants of valproic acid, along with their specific genetic polymorphism and implications [17].
The ABCB1 gene, encoding P-glycoprotein, influences the efflux of several ASMs from the central nervous system. Polymorphisms such as 3435C>T have been associated with varying drug transportation activity, leading to differences in drug response among patients with epilepsy. This particularly impacts drugs like carbamazepine (CBZ) and lamotrigine [18]. A comprehensive analysis encompassing 18 studies was conducted with 3,293 epilepsy patients, revealing the significant role of ABCB1 c.3435C>T polymorphism in CBZ metabolism and resistance. It was found that individuals carrying the T allele had adjusted concentrations of CBZ significantly different from those with the C allele. This indicated that this polymorphism directly affects CBZ plasma levels and in turn, its efficacy and side-effect profile. Moreover, the EPHX1 c.416A>G polymorphism was linked to the metabolism of CBZ, specifically its conversion to CBZ-10,11-trans-dihydrodiol (CBZD). These findings emphasize the complex genetic factors that influence ASM metabolism and resistance, highlighting the need for a more personalized approach to epilepsy treatment that takes into account individual genetic profiles (Table 2) [19].
Genetic polymorphism and drug transporters
| Drug transporters | Genetic polymorphisms | Implications | Reference(s) |
|---|---|---|---|
| ABCB1 (P-glycoprotein) | 3435C>T | Associated with varying drug transportation activity, impacting drug response for carbamazepine and lamotrigine. Significant role in carbamazepine metabolism and resistance; T allele carriers show significantly different carbamazepine plasma levels. Influence on anticonvulsant drug efflux from brain tissue, potentially contributing to drug resistance; CC genotype associated with higher efflux. | [18–21] |
| EPHX1 | c.416A>G | Linked to carbamazepine metabolism, specifically conversion to carbamazepine-10,11-trans-dihydrodiol (CBZD). | [19] |
Studies further elaborated on the relationship between ABCB1 polymorphisms and CBZ metabolism through a meta-analysis of 12 studies involving 2,126 epilepsy patients. This analysis highlighted the TC genotype of the rs1045642 polymorphism as significantly associated with decreased CBZ plasma concentration, suggesting an alteration in the drug’s efflux mechanism that could impact its therapeutic efficacy. It was also found that the AG genotype of the rs2032582 polymorphism correlated with increased CBZ concentration, pointing towards genetic variants that modulate the pharmacokinetics of CBZ integration of pharmacogenetic testing in the clinical management of epilepsy to achieve optimal treatment outcomes (Figure 2) [20].
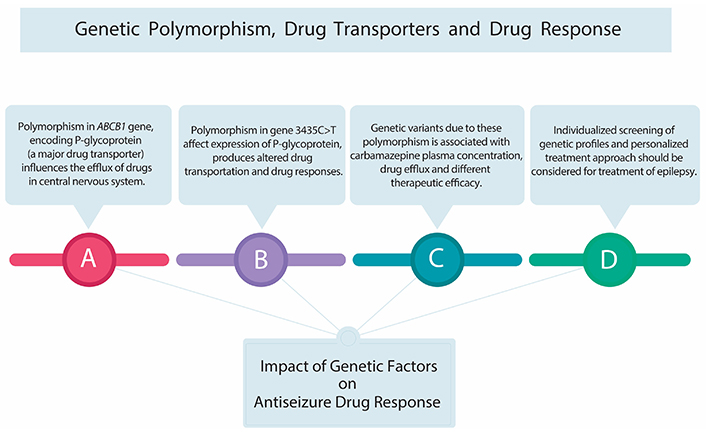
Genetic polymorphism, drug transporter, and drug response. This figure highlights genetic influences on drug response, emphasizing P-glycoprotein’s role as a vital drug transporter. Polymorphisms in P-glycoprotein can affect carbamazepine metabolism, requiring dose adjustments and genetic testing for safe and effective antiseizure medication use. Notably, the 3435C>T variant of the ABCB1 gene influences P-glycoprotein expression, impacting drug response and necessitating consideration of individual genetic makeup
A study investigated the influence of the ABCB1 C3435T polymorphism on anticonvulsant drug resistance. The study emphasized the role of the gene in regulating drug efflux across the blood-brain barrier. The meta-analysis suggested that individuals with the 3435CC genotype may experience a higher efflux of anticonvulsants from brain tissue, which could contribute to pharmaco-resistant. This finding highlights the importance of genetic testing in identifying patients at risk of DRE [21].
A multicenter cohort study and meta-analysis were conducted to delve into the role of polymorphisms in SCN1A, SCN2A, and SCN3A genes, which encode voltage-gated sodium channels, a common target for ASMs. Despite examining 39 polymorphisms across two epilepsy cohorts from Malaysia and Hong Kong, encompassing 1,504 patients, and conducting a meta-analysis on specific polymorphisms (SCN1A rs3812718 and rs2298771, and SCN2A rs17183814), the study found no significant association of these polymorphisms with ASM responsiveness. This comprehensive investigation suggests that common variants in these genes do not majorly influence ASM response, highlighting the complexity of epilepsy pharmacogenetics and the need for exploring beyond these variants to understand drug resistance and efficacy [22].
A meta-analysis was conducted to focused on SCN1A rs2298771, SCN1A rs10188577, SCN2A rs17183814, and SCN2A rs2304016 polymorphisms revealing a significant association of SCN1A rs2298771 with ASM resistance. Analysis of 19 studies confirmed that carriers of the G allele in SCN1A rs2298771 have an increased risk of ASM resistance, especially in Caucasians from South Asia. However, no significant associations were found for other studied polymorphisms, indicating the specificity of SCN1A rs2298771’s impact on ASM resistance and highlighting the importance of genetic background in epilepsy treatment [23].
In 2023, Khatri and colleagues [24] conducted a study that used an in silico-based approach to identify new targets associated with the SCN1A gene that could potentially be used to develop ASM with minimal side effects and maximum efficacy. The study examined 157 genes related to epilepsy and, through network analysis, identified proteins encoded by the SCN9A, HCN2, and FGF12 genes as potential targets. This investigation offers a unique perspective on identifying targets for ASM and highlights the potential of using genetic and network analyses to uncover new treatment options for epilepsy [24].
Newer medications such as levetiracetam, lacosamide, and perampanel are being researched in addition to the already mentioned ASMs. For instance, genetic factors that influence SV2A (the synaptic vesicle glycoprotein targeted by levetiracetam) and collagen-related genes that impact lacosamide’s modulation of sodium channels provide new possibilities for personalized epilepsy therapy [25].
Approximately 30% of epilepsy patients show pharmaco-resistance despite adequate therapy using ASM. Figure 3 clearly illustrates the investigations that show impacts of genetic determinants on drug resistance. Identification of a null allele of CYP2C9 in an African-American exhibiting toxicity to phenytoin [8]. Genetic investigations predominantly centered on drug transporters’ role at the barrier between blood and brain, as substantial proof implies that heightened functional activity of multidrug transport proteins may impede the entry of ASMs into the brain. Therefore, genetic polymorphisms might cause a heightened expression of these drug transporters, contributing to ASM resistance.
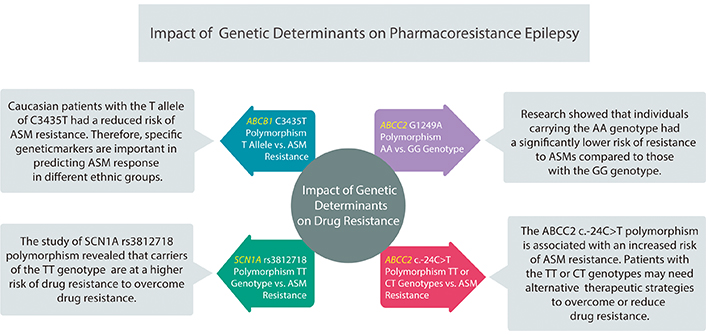
Impact of genetic determinants on pharmaco-resistant epilepsy. Genetic factors significantly influence antiseizure medication (ASM) resistance, necessitating personalized treatment considering genetic variations and ethnic diversity. It was found that the ABCC2 AA genotype carriers had lower resistance risk, while ABCB1 C3435T T allele reduced resistance risk in Caucasians. Conversely, SCN1A rs3812718 TT carriers showed higher resistance risk
The ABCC2 gene plays an important role in drug transport. A research study by Chen et al. [26] conducted on 2,213 patients examined a particular genetic variation in ABCC2 called G1249A polymorphism. The findings of their research showed that individuals carrying the AA genotype had a significantly lower risk of resistance to ASMs compared to those with the GG genotype. The odds ratio (OR) was 0.372 with a 95% confidence interval (CI) of 0.182–0.762, indicating a protective effect against pharmaco-resistance. This relationship was more pronounced under a recessive model where the OR was 0.399 with a 95% CI of 0.200–0.795. These findings underscore the potential of genetic screening in personalizing ASM selection for better treatment outcomes. Therefore, understanding genetic predispositions such as ABCC2 G1249A polymorphism is crucial in clinical practice [26].
Moreover, research on ABCB1 polymorphisms found that Caucasian patients with the T allele of C3435T had a reduced risk of ASM resistance. This suggests that specific genetic markers are important in predicting ASM response in different ethnic groups. These findings provide new opportunities for ethnic-specific genetic testing, which can enhance the effectiveness of ASM treatment [27]. The ABCC2 c.-24C>T polymorphism is associated with an increased risk of ASM resistance. This suggests that the polymorphism could be a potential predictor for pharmaco-resistance, and patients with the TT or CT genotypes may need alternative therapeutic strategies to overcome or reduce drug resistance [28].
Similarly, the study of SCN1A rs3812718 polymorphism revealed that carriers of the TT genotype were at a higher risk of drug resistance highlighting the clinical significance of genetic testing in pediatric epilepsy to predict valproic acid response personalized treatment accordingly [29].
Personalized medicine has increasingly influenced the management of epilepsy. A deeper understanding of epilepsy genetics, coupled with advancements in in vivo and in vitro genetic frameworks, has propelled the use of genetic approaches in devising targeted therapies. Identifying genetic factors underlying epilepsy has significantly expanded our knowledge of epileptogenic mechanisms. However, achieving positive outcomes through this approach is complex. The interaction of genetic variants within the entirety of the genome and the presence of adaptive and compensatory changes can render the direct correction of the identified dysfunction challenging. This highlights a critical consideration in precision medicine. Therefore, treatments must target the mechanisms directly associated with causative genetic alterations.
Recent investigations into precision medicine therapies have provided promising directions. For instance, patients with GLUT1 deficiency have shown remarkable improvement on a ketogenic diet, which addresses the transport dysfunction by providing an alternative energy source through ketogenesis, thereby managing seizures and paroxysmal activities. Similarly, pyridoxine-dependent epilepsy, linked to antiquities deficiency, responds well to vitamin B6 therapy, showing the direct impact of tailored nutrient supplementation on genetic epilepsies. Moreover, the application of sodium channel blockers in cases with KCNQ2, SCN1A, and SCN2A mutations, and the use of mTOR-inhibitors in mTORopathies, exemplify the repurposing of available treatments based on genetic insights as shown in Table 3 [30].
Genetic disorders and relative approach of precision medicine
| Genetic disorder | Approach of precision medicine | Reference |
|---|---|---|
| GLUT1 deficiency | Ketogenic diet addresses the transport dysfunction and managing seizures and paroxysmal activities | [30] |
| Pyridoxine-dependent epilepsy | Direct impact on genetic epilepsies is shown on vitamin B6 therapies | [30] |
| KCNQ2, SCN1A, SCN2A mutations | Sodium channel blockers applied for certain mutations | [30] |
| mTORopathies | mTOR inhibitors used as targeted therapy | [30] |
| Exome and genome sequencing | Enhanced genotype-phenotype correlations, guiding pyridoxine supplementation | [31] |
| KCNT1-related epileptic disorders | Quinidine use as genotype-directed therapy | [32] |
| KCNQ2-related epilepsy | Analysis of treatment approaches for precision medicine | [33] |
Several studies have highlighted the role of advanced diagnostic tools, such as exome and genome sequencing, in enhancing genotype-phenotype correlations. These findings support the use of pyridoxine supplementation in managing seizures across various genetic epilepsies and suggest ganaxolone as a novel therapeutic agent for certain syndromes, signaling a leap toward personalized treatment modalities [31]. The quinidine uses in KCNT1-related epileptic disorders and analysis of treatment approaches in KCNQ2-related epilepsy further elaborate the precision medicine in epilepsy. These studies illustrate the potential of genotype-directed therapies, highlighting the crucial balance between efficacy and safety in the development of new treatments [32, 33] also summarized in Table 3.
The safety of ASMs is significantly influenced by genetic variants, with different individuals experiencing severe hypersensitivity responses such as TEN and SJS, potentially leading to fatal effects. Key developments in pharmacogenomics have illuminated the role of specific genetic variants in predicting and mitigating these risks. The 1,502 gene variants of HLA-B have been identified as a significant marker for severe cutaneous adverse reactions during CBZ therapy, especially within Asian populations. There is a strong association between HLA-B*1502 and CBZ-induced SJS and TEN, particularly among Han-Chinese, Thai, and Malaysian populations, emphasizing the necessity for genetic screening before CBZ treatment in these communities to prevent TEN and SJS [34] as shown in Table 4. Conversely, in European populations, the HLA-A*3101 allele has been associated with an increased susceptibility to CBZ-induced hypersensitivity reactions. HLA-A*3101 is a universal risk marker for CBZ-induced cutaneous adverse drug reactions, irrespective of the clinical outcome, underscoring the global relevance of pharmacogenomic testing for ASM safety [35].
Genetic variants associated with ASM safety
Pharmacogenomics holds immense promise in revolutionizing the management of DRE by personalizing treatment approaches based on individual genetic profiles. Through the identification of genetic variations influencing drug response, clinicians can tailor antiepileptic drug therapy, maximizing efficacy while minimizing adverse effects. Embracing and integrating these advancements into clinical practice can lead to a better quality of life and renewed hope for individuals with DRE.
ASM: antiseizure medication
CBZ: carbamazepine
DRE: drug-resistant epilepsy
SJS: Stevens-Johnson syndrome
TEN: toxic epidermal necrolysis
AS: Conceptualization, Writing—original draft. KH: Writing—original draft, Writing—review & editing. A Zafar: Validation, Writing—review & editing. A Zainab: Conceptualization, Writing—original draft, Writing—review & editing. SA: Conceptualization, Supervision. All authors read and approved the submitted version.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
© The Author(s) 2024.
Copyright: © The Author(s) 2024. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
