Abstract
Pregnancy complications such as pre-eclampsia, gestational diabetes, and intrauterine growth restriction (IUGR) not only present immediate risks to maternal and fetal health but also have long-term implications for the cardiovascular health of offspring. Emerging evidence suggests that these complications may induce epigenetic changes, which in turn predispose offspring to cardiovascular diseases (CVDs) later in life. Epigenetic modifications, including DNA methylation, histone modifications, and non-coding RNA regulation, play crucial roles in fetal development by influencing gene expression without altering the DNA sequence. Aberrant DNA methylation patterns have been observed in offspring exposed to adverse intrauterine environments, affecting genes that regulate blood pressure, lipid metabolism, and inflammation, key factors in CVDs development. Similarly, histone modifications linked to pregnancy complications can disrupt the expression of genes involved in vascular function, contributing to increased cardiovascular risk. Additionally, dysregulation of microRNAs in response to complications like gestational diabetes may influence pathways related to insulin signaling and atherosclerosis. This review synthesizes current knowledge on the epigenetic mechanisms by which pregnancy complications increase CVDs risk in offspring, highlighting potential avenues for early intervention and therapeutic strategies. Understanding these mechanisms could lead to the development of targeted interventions during pregnancy, potentially reducing the intergenerational transmission of cardiovascular risk and improving long-term health outcomes for both mothers and their children.
Keywords
CVDs, DNA methylation, epigenetics, histone modification, microRNAsIntroduction
Pregnancy is a critical period that profoundly impacts both maternal and fetal health, with potential long-term consequences for the offspring [1]. While the immediate risks associated with pregnancy complications such as preeclampsia, gestational diabetes mellitus (GDM), and intrauterine growth restriction (IUGR) are well recognized, an emerging body of research suggests that these complications also have significant long-term implications, particularly concerning the cardiovascular health of the offspring [2]. The concept that adverse intrauterine environments can predispose individuals to chronic diseases later in life, including cardiovascular diseases (CVDs), has been increasingly supported by studies in developmental origins of health and disease (DOHaD) [3]. Central to this concept is the role of epigenetic mechanisms, which refer to heritable changes in gene expression that occur without alterations in the underlying DNA sequence [4]. These mechanisms are vital in regulating gene activity during fetal development, and disruptions in normal epigenetic programming can lead to lasting alterations in gene expression that may predispose individuals to various health conditions, including CVDs [5]. Epigenetic modifications encompass several processes, including DNA methylation, histone modification, and regulation by non-coding RNAs (ncRNAs), each of which plays a crucial role in the developmental processes that occur during pregnancy [6]. DNA methylation involves the addition of a methyl group to the cytosine residues in DNA, typically resulting in the suppression of gene expression. This process is essential for normal development, and aberrations in DNA methylation patterns have been linked to various pregnancy complications (Table 1). For example, in pre-eclampsia a condition characterized by high blood pressure and damage to organ systems, particularly the liver and kidneys altered DNA methylation patterns have been observed in both the placenta and fetal tissues [17]. These changes often affect genes involved in vascular development, inflammatory responses, and metabolic regulation, all of which are critical for maintaining cardiovascular health. In offspring, exposure to such altered methylation patterns during critical periods of development can lead to long-term dysregulation of genes that control essential functions like blood pressure, lipid metabolism, and glucose homeostasis [18]. These dysregulations can manifest as increased susceptibility to hypertension, dyslipidemia, and chronic inflammation, all of which are significant risk factors for the development of CVDs. Previous research suggests that the epigenetic mechanisms linking pregnancy complications to CVD in offspring represent a complex and multifaceted area of research that holds significant promise for improving our understanding of the DOHaD [19]. By elucidating the specific epigenetic pathways involved, we can gain insights into how adverse intrauterine environments contribute to long-term health outcomes, and potentially develop targeted interventions to reduce the intergenerational transmission of cardiovascular risk [20]. This not only has implications for improving the health of future generations but also underscores the importance of maternal health and well-being during pregnancy as a critical determinant of long-term health outcomes for both mothers and their children [21]. As research in this area continues to evolve, our understanding of the epigenetic links between pregnancy complications and CVD will likely deepen, leading to new strategies for preventing and managing cardiovascular risk from the earliest stages of life [22]. This review explores the epigenetic pathways through which pregnancy complications may increase the risk of CVD in offspring.
Detailed epigenetic mechanisms linking pregnancy complications to CVD in offspring
| Epigenetic mechanism | Pregnancy complications | Key genes/pathways affected | Potential cardiovascular outcomes in offspring | References |
|---|---|---|---|---|
| DNA methylation | Preeclampsia | ACE, AGTR1, IL-6, PPAR | Hypertension, dyslipidemia, chronic inflammation | [7] |
| DNA methylation | GDM | INS, PPARGC1A, ADIPOQ | Metabolic syndrome, insulin resistance, atherosclerosis | [8] |
| DNA methylation | IUGR | VEGF, NOS3, HIF1A | Increased vascular stiffness, hypertension, atherosclerosis | [9] |
| Histone modifications | Preeclampsia | H3K27me3 (polycomb repressive complex 2), H4K8ac | Altered gene expression in vascular development, increased risk of hypertension | [10] |
| Histone modifications | IUGR | H3K9me3, H4K20me3 | Vascular dysfunction increases cardiovascular risk | [11] |
| Non-coding RNAs (miRNAs) | GDM | miR-29, miR-143, miR-155 | Increased risk of insulin resistance, atherosclerosis, hypertension | [12] |
| Non-coding RNAs (miRNAs) | Preeclampsia | miR-210, miR-126, miR-155 | Increased cardiovascular risk, endothelial dysfunction | [13] |
| Chromatin remodeling | Preeclampsia, IUGR | SMARCA4, CHD4, BRG1 | Altered gene expression in cardiovascular development, increased risk of CVDs | [14] |
| Long non-coding RNAs (lncRNAs) | GDM, IUGR | H19, MEG3, XIST | Altered metabolic regulation, increased risk of CVDs | [15] |
| RNA editing | GDM, IUGR | ADARB1, ADARB2, APOBEC1 | Altered RNA processing, increased cardiovascular risk | [16] |
GDM: gestational diabetes mellitus; IUGR: intrauterine growth restriction; ACE: angiotensin-converting enzyme; AGTR1: angiotensin II receptor type 1; PPAR: peroxisome proliferator-activated receptor; INS: insulin; ADIPOQ: adiponectin; PPARGC1A: peroxisome proliferator-activated receptor gamma, coactivator 1 alpha; VEGF: vascular endothelial growth factor; NOS3: nitric oxide synthase 3, endothelial; HIF1A: hypoxia inducible factor 1 alpha subunit; SMARCA4: SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4; CHD4: chromodomain helicase DNA binding protein 4; BRG1: brahma-related gene 1; MEG3: maternally expressed gene 3; XIST: X-inactive specific transcript; ADARB1: adenosine deaminase RNA-specific B1; APOBEC1: apolipoprotein b editing catalytic subunit; CVD: cardiovascular disease
Epigenetic modifications and CVD
Epigenetic modifications include DNA methylation, histone modification, and the regulation of gene expression by ncRNAs. These mechanisms are crucial in regulating gene expression during development, and they can be influenced by environmental factors, including those encountered during pregnancy [11]. Disruptions in normal epigenetic programming during fetal development can have lasting effects on gene expression patterns, potentially leading to altered physiological functions and increased disease susceptibility (Figure 1) [23].
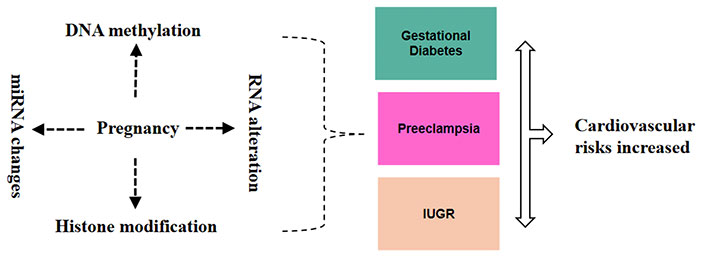
Epigenetic and RNA alterations during pregnancy and their link to cardiovascular risk. These alterations such as DNA methylation, RNA modification, histone modification, and miRNA changes during pregnancy can lead to conditions like gestational diabetes mellitus (GDM), preeclampsia, and intrauterine growth restriction (IUGR), which are linked to increased cardiovascular risks for both the mother and the fetus
DNA methylation
DNA methylation is a fundamental epigenetic mechanism involving the addition of a methyl group to the 5-position of cytosine residues within CpG dinucleotides [9]. This modification typically leads to the repression of gene expression by altering the accessibility of the DNA to transcriptional machinery [18]. In the context of pregnancy, DNA methylation patterns are crucial for regulating gene expression during fetal development, and disruptions in these patterns can have significant long-term implications for health. Pregnancy complications such as pre-eclampsia, GDM, and IUGR have been associated with altered DNA methylation patterns in both maternal and fetal tissues [9]. These modifications can influence the expression of genes involved in cardiovascular development and function, potentially predisposing offspring to CVDs later in life [24]. Preeclampsia, a pregnancy complication characterized by hypertension and multi-organ dysfunction, is associated with significant alterations in DNA methylation patterns. Studies have demonstrated that preeclampsia leads to aberrant DNA methylation in genes involved in vascular development, inflammatory responses, and placental function [17]. For instance, increased methylation of the angiotensin-converting enzyme (ACE) gene has been linked to altered blood pressure regulation in offspring. Similarly, hypermethylation of angiotensin II receptor type 1 (AGTR1) in the placenta can affect the development of the fetal cardiovascular system, leading to increased susceptibility to hypertension in later life [25]. Furthermore, differential methylation of genes related to oxidative stress and inflammation, such as nuclear factor kappa b subunit 1 (NF-KB1), has been observed in the context of preeclampsia. This dysregulation may contribute to chronic inflammation and endothelial dysfunction, which are key factors in the development of CVDs [26]. GDM characterized by glucose intolerance during pregnancy, also affects DNA methylation patterns. Aberrant methylation of genes involved in glucose metabolism and lipid homeostasis has been observed in the offspring of mothers with GDM [27]. For example, hypomethylation of the peroxisome proliferator-activated receptor gamma (PPAR-γ) gene has been linked to dysregulated lipid metabolism and increased risk of metabolic syndrome and CVDs in offspring [28]. Additionally, changes in DNA methylation of Leptin (LEP) and adiponectin (ADIPOQ) genes, which play roles in energy regulation and insulin sensitivity, further illustrate how GDM-induced methylation alterations can predispose offspring to cardiovascular and metabolic disorders [29].
IUGR, resulting from placental insufficiency, leads to altered DNA methylation patterns in the fetus that can influence cardiovascular health later in life. Aberrant methylation of genes such as insulin-like growth factor 2 (IGF2) and 11β-hydroxysteroid dehydrogenase type 2 (HSD11B2) has been observed in IUGR conditions [30]. IGF2, a key regulator of growth and development, exhibits differential methylation that can impact fetal growth and development, while alterations in HSD11B2 can affect glucocorticoid metabolism, influencing cardiovascular outcomes [31]. Studies have shown that IUGR-related changes in DNA methylation can lead to long-term effects on blood pressure regulation, glucose metabolism, and endothelial function. These alterations may predispose individuals to hypertension, insulin resistance, and CVDs later in life [32]. The epigenetic changes induced by pregnancy complications can have lasting effects on cardiovascular health through several mechanisms. Altered DNA methylation can lead to persistent gene expression changes that affect vascular development, metabolic regulation, and inflammatory responses. For example, genes involved in endothelial function and vascular remodeling may remain dysregulated due to aberrant methylation, leading to an increased risk of CVDs such as atherosclerosis and hypertension [33]. Moreover, the inheritance of these epigenetic marks can influence not only the immediate offspring but also subsequent generations, potentially contributing to a cycle of cardiovascular risk. Understanding these mechanisms provides valuable insights into how early-life exposures shape long-term health outcomes and highlights the importance of early intervention to mitigate these risks [34]. DNA methylation, the addition of a methyl group to the cytosine residues in DNA, is one of the most studied epigenetic modifications. CpG islands, which are methylation-susceptible regions commonly found near gene promoters, play a significant role in gene expression regulation. These regions are often subject to DNA methylation, influencing transcriptional activity [35]. Aberrant DNA methylation patterns have been associated with various pregnancy complications. For instance, in cases of preeclampsia, there is evidence of altered DNA methylation in both the placenta and fetal tissues [36]. These changes can affect genes involved in vascular development, inflammatory responses, and metabolic regulation, all of which are critical for cardiovascular health. In offspring, altered DNA methylation patterns due to exposure to adverse intrauterine environments can lead to dysregulation of genes that control blood pressure, lipid metabolism, and glucose homeostasis key factors in the development of CVDs [37]. Studies have shown that children born to mothers with preeclampsia have differentially methylated regions in genes associated with hypertension and heart disease, suggesting a mechanistic link between maternal complications and offspring cardiovascular risk [38]. Recent studies have shown that placental DNA methylation changes are closely linked to various pregnancy complications and may influence long-term health [39]. For instance, increased global placental DNA methylation has been associated with elevated blood pressure during pregnancy, and with conditions like being born large for gestational age [40]. Additionally, abnormal DNA methylation of the glucocorticoid receptor gene promoter in the placenta has been linked to blood pressure regulation. Similarly, increased placental DNA methylation levels are associated with gestational diabetes. These alterations can impact critical developmental processes, potentially leading to long-term cardiovascular risks such as hypertension and dyslipidemia in offspring [41].
Histone modifications
Histone proteins, around which DNA is wrapped, undergo various post-translational modifications, such as acetylation, methylation, and phosphorylation, which can influence gene expression by altering chromatin structure. Histone modifications represent another crucial aspect of epigenetic regulation, involving post-translational changes to histone proteins that can influence gene expression by altering chromatin structure. These modifications include acetylation, methylation, phosphorylation, and ubiquitination, among others [10]. Histone modifications play a crucial role in regulating gene expression through changes in chromatin structure. These modifications, including acetylation, methylation, and phosphorylation, can profoundly influence cardiovascular risk by altering the transcriptional activity of genes involved in vascular function and inflammation [42].
Acetylation and cardiovascular risk: histone acetylation, which is generally associated with gene activation, affects several genes related to cardiovascular health [43]. For instance, increased acetylation of histone H3 at the promoter regions of pro-inflammatory cytokine genes, such as TNF-α and IL-6, has been linked to elevated inflammation and endothelial dysfunction, both of which are risk factors for CVDs [44]. Studies have shown that the histone acetyltransferase (HAT) p300/CBP plays a significant role in promoting the expression of these inflammatory cytokines in response to oxidative stress, thereby contributing to atherosclerosis development [43].
Methylation and cardiovascular risk: histone methylation can either activate or repress gene expression, depending on the specific residues modified [45]. For example, trimethylation of histone H3 at lysine 4 (H3K4me3) is associated with gene activation, whereas H3K27me3 is linked to gene repression. In the context of cardiovascular risk, aberrant methylation patterns have been observed in genes associated with vascular remodeling and hypertension [46]. A notable study highlighted that reduced levels of H3K4me3 in the promoter regions of the endothelin-1 (ET-1) gene are associated with increased ET-1 expression, contributing to vasoconstriction and elevated blood pressure [47].
Phosphorylation and cardiovascular risk: histone phosphorylation, particularly at serine 10 and threonine 3, is implicated in the regulation of gene expression in response to stress [48]. Increased phosphorylation of histone H3 at serine 10 has been observed in response to hypertensive stimuli, which can lead to the activation of genes involved in cardiac hypertrophy and fibrosis [49]. For instance, research has shown that histone H3 phosphorylation is involved in the transcriptional regulation of genes such as atrial natriuretic peptide (ANP) and brain natriuretic peptide (BNP), which are markers of cardiac stress and remodeling [48].
Clinical relevance and therapeutic potential: understanding the specific impacts of these histone modifications can inform the development of targeted epigenetic therapies. For example, inhibitors of specific HATs or histone methyltransferases may offer therapeutic potential in modulating gene expression related to cardiovascular risk factors. Clinical trials are beginning to explore such strategies, aiming to refine the use of epigenetic drugs in cardiovascular medicine.
The balance and pattern of these all-histone modifications are critical for the regulation of gene expression during development. Pregnancy complications like IUGR have been associated with aberrant histone modifications in the fetus. IUGR, often resulting from placental insufficiency, exposes the fetus to a suboptimal intrauterine environment, which can lead to altered histone acetylation at specific gene loci [50]. These modifications can persist long after birth, leading to sustained alterations in gene expression that predispose the offspring to CVDs. For instance, changes in histone acetylation and methylation at genes involved in endothelial function and vascular smooth muscle cell proliferation can result in increased vascular stiffness, hypertension, and atherosclerosis in later life [51]. Studies in animal models have provided compelling evidence that maternal undernutrition a common cause of IUGR leads to specific histone modifications that contribute to cardiovascular dysfunction in the offspring, underscoring the long-term impact of epigenetic changes induced by adverse intrauterine conditions [52]. The implications of these epigenetic changes are profound, as they suggest that the conditions experienced in utero can have lasting effects on an individual’s health, potentially setting the stage for the development of chronic diseases such as CVDs many years or even decades later. This highlights the importance of understanding the epigenetic mechanisms at play, not only to elucidate the pathways through which pregnancy complications exert their long-term effects but also to identify potential points of intervention [9]. If we can understand how these epigenetic changes are initiated and maintained, it may be possible to develop strategies to mitigate their effects, either during pregnancy or in the early postnatal period. For instance, targeted nutritional interventions or pharmacological treatments during pregnancy could potentially reverse or ameliorate adverse epigenetic modifications, reducing the risk of CVDs in the offspring. Additionally, advances in epigenetic editing technologies, such as CRISPR-based systems, offer the possibility of precisely targeting and correcting aberrant epigenetic marks. However, these approaches are still in their infancy, and significant challenges remain in terms of ensuring their safety, efficacy, and ethical acceptability [53].
ncRNAs
ncRNAs are a diverse group of RNA molecules that do not encode proteins but play crucial roles in regulating gene expression at various levels [54]. Among the ncRNAs, microRNAs (miRNAs) are particularly important in the context of pregnancy complications and their long-term effects on offspring health. miRNAs are short, single-stranded RNAs that typically regulate gene expression post-transcriptionally by binding to complementary sequences on messenger RNAs (mRNAs), leading to their degradation or inhibition of translation [55]. The dysregulation of miRNAs has been implicated in numerous pathological conditions, including CVDs, and can be influenced by adverse pregnancy conditions. Pregnancy complications such as pre-eclampsia, GDM, and maternal obesity have been shown to alter miRNA expression profiles in both maternal and fetal tissues [56]. These changes can have significant implications for cardiovascular health in offspring, influencing gene networks involved in metabolic regulation, inflammation, and vascular function. Preeclampsia, a condition characterized by high blood pressure and damage to organs, significantly affects miRNA expression. Several miRNAs have been found to be dysregulated in preeclampsia, which may contribute to the increased cardiovascular risk observed in offspring. For example, miR-210 is upregulated in pre-eclampsia and plays a role in the regulation of angiogenesis and hypoxic responses. In the context of preeclampsia, miR-210 contributes to impaired placental blood flow and endothelial dysfunction, which can impact fetal cardiovascular health [57]. Additionally, miR-126, which is involved in endothelial cell function and vascular integrity, is downregulated in preeclampsia. This downregulation can lead to disrupted endothelial function and increased risk of vascular diseases in the offspring. The altered expression of these miRNAs suggests a mechanism by which pre-eclampsia-induced changes in miRNA profiles can have long-term effects on cardiovascular health [58]. GDM is associated with significant changes in miRNA expression that can influence cardiovascular risk in offspring. For instance, miR-29 and miR-143 are known to regulate insulin signaling and lipid metabolism. In the context of GDM, the dysregulation of these miRNAs has been linked to impaired glucose metabolism and increased risk of metabolic syndrome in the offspring. miR-29 is downregulated in GDM, which can affect insulin signaling pathways and contribute to insulin resistance and increased cardiovascular risk [59]. miR-143, which regulates vascular smooth muscle cell function and is involved in the development of atherosclerosis, is also altered in response to GDM. The disruption of miR-143 expression can lead to changes in vascular function and contribute to the development of CVDs. These findings indicate that GDM-induced miRNA alterations can have lasting effects on cardiovascular health through their impact on glucose and lipid metabolism, as well as vascular function [15]. Maternal obesity is another pregnancy complication associated with altered miRNA expression, which can influence cardiovascular risk in offspring. miR-122 and miR-33 are key miRNAs involved in lipid metabolism and cholesterol homeostasis. In maternal obesity, the expression of miR-122 is altered, leading to dysregulated lipid metabolism and an increased risk of CVDs in offspring. miR-33 also plays a role in regulating cholesterol homeostasis, and its dysregulation in maternal obesity has been linked to altered cholesterol levels and increased cardiovascular risk [60]. The impact of maternal obesity on miRNA profiles highlights the importance of managing maternal weight to prevent adverse cardiovascular outcomes in offspring. Dysregulation of these miRNAs can lead to lasting changes in lipid metabolism and cardiovascular health, further emphasizing the need for early interventions. The dysregulation of miRNAs due to pregnancy complications can have profound implications for cardiovascular health by influencing gene networks involved in metabolism, inflammation, and vascular function. miRNAs act as fine tuners of gene expression, and their alteration can lead to sustained changes in the expression of genes critical for cardiovascular health [61]. For instance, miRNA-mediated regulation of genes involved in endothelial function, insulin signaling, and lipid metabolism can have long-term effects on the development of CVDs. Moreover, the inheritance of altered miRNA expression profiles can potentially affect subsequent generations, leading to a cycle of increased cardiovascular risk. Understanding the mechanisms by which pregnancy complications influence miRNA expression and their impact on cardiovascular health is crucial for developing strategies to mitigate these risks [62]. Understanding the epigenetic mechanisms that link pregnancy complications to CVDs in offspring opens the door to potential interventions. Early identification of at-risk pregnancies and targeted epigenetic therapies could mitigate the adverse outcomes for offspring. For example, nutritional interventions, pharmacological agents, or lifestyle modifications during pregnancy might beneficially influence epigenetic marks, reducing the risk of CVDs in the next generation [63]. Furthermore, advancements in epigenetic editing technologies, such as CRISPR-based systems, offer promising avenues for precisely targeting and modifying aberrant epigenetic marks. However, the ethical and safety considerations of such approaches need careful evaluation before clinical application [61].
Clinical implications of epigenetic findings and early identification of epigenetic markers
Prenatal screening: the identification of specific epigenetic markers linked to cardiovascular risks could be integrated into prenatal screening programs. For example, detecting DNA methylation patterns associated with adverse intrauterine environments in early pregnancy could help in assessing the risk of future cardiovascular conditions in offspring [64].
Risk stratification: epigenetic markers could be used to stratify risk in pregnant women. By identifying those with high-risk epigenetic profiles, personalized prenatal care plans could be developed to mitigate potential risks through lifestyle modifications, nutritional interventions, or increased monitoring [65].
Epigenetic drug development: insights into the specific epigenetic modifications involved in cardiovascular risk could lead to the development of targeted therapies. For instance, drugs that modify histone acetylation or methylation patterns could be developed to counteract the detrimental effects of epigenetic changes observed in CVDs [66].
Gene editing technologies: advanced technologies like CRISPR/Cas9 could be employed to correct specific epigenetic alterations. For example, editing histone modifications or DNA methylation patterns in targeted tissues could potentially reverse abnormal gene expression associated with cardiovascular risk [67].
Integration into clinical practice
Personalized medicine: epigenetic insights could enhance personalized medicine approaches by tailoring interventions based on an individual’s epigenetic profile. This includes customizing treatment plans and preventive strategies for those identified as having a higher genetic risk of CVDs [68].
Clinical trials: clinical trials should focus on evaluating the efficacy of epigenetic therapies in preventing or treating CVDs. Data from these trials would be crucial in determining the safety and effectiveness of new treatments and integrating them into standard care practices [22].
Ethical and practical considerations
The use of epigenetic modifications and gene editing in clinical settings must be accompanied by ethical considerations [69]. Issues related to consent, potential long-term effects, and equity of access to these advanced treatments need to be addressed. Moreover, developing and implementing epigenetic interventions may involve significant costs. Ensuring that these interventions are accessible to a broad population and are integrated into existing healthcare systems is essential for maximizing their benefit [70].
Conclusions
The epigenetic mechanisms connecting pregnancy complications to CVDs in offspring are intricate and continually developing. Key epigenetic factors, including DNA methylation, histone modifications, and ncRNAs, play critical roles in mediating the long-term cardiovascular risks associated with adverse intrauterine environments. As research progresses, it may become feasible to develop targeted interventions to mitigate the intergenerational transmission of CVDs risk, thereby enhancing health outcomes for future generations. To advance this field, future research should prioritize several key areas including the longitudinal study which means conducting comprehensive longitudinal studies to track the long-term cardiovascular effects of epigenetic modifications from prenatal environments into adulthood. This will provide valuable insights into the persistence and impact of these modifications over time. Another major step is targeted Interventions which explore and evaluate specific interventions aimed at correcting detrimental epigenetic modifications. Research should focus on developing and testing therapies that can reverse or modulate epigenetic changes linked to CVDs risk. Furthermore, advanced Epigenetic Technologies utilize cutting-edge technologies for precise epigenetic editing, such as CRISPR-based approaches, to refine and test potential therapeutic strategies. These tools will enhance our ability to understand and manipulate epigenetic mechanisms with high precision.
Abbreviations
| CVDs: | cardiovascular diseases |
| DOHaD: | developmental origins of health and disease |
| ET-1: | endothelin-1 |
| GDM: | gestational diabetes mellitus |
| H3K4me3: | trimethylation of histone H3 at lysine 4 |
| HAT: | histone acetyltransferase |
| HSD11B2: | 11β-hydroxysteroid dehydrogenase type 2 |
| IGF2: | insulin-like growth factor 2 |
| IUGR: | intrauterine growth restriction |
| miRNAs: | microRNAs |
| ncRNAs: | non-coding RNAs |
Declarations
Author contributions
KRT: Conceptualization, Methodology, Supervision. IB: Writing—review & editing. MWA: Formal analysis, Writing—review & editing.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent to publication
Not applicable.
Availability of data and materials
Not applicable.
Funding
Not applicable.
Copyright
© The Author(s) 2024.