Affiliation:
1Optiscan Imaging Ltd, Melbourne 3170, Australia
2Australian Centre for Oral Oncology Research & Education, Perth 6009, Australia
ORCID: https://orcid.org/0000-0003-2042-1809
Affiliation:
1Optiscan Imaging Ltd, Melbourne 3170, Australia
2Australian Centre for Oral Oncology Research & Education, Perth 6009, Australia
Email: ceo@optiscan.com
ORCID: https://orcid.org/0000-0002-1642-6204
Explor Digit Health Technol. 2025;3:101138 DOI: https://doi.org/10.37349/edht.2025.101138
Received: September 10, 2024 Accepted: November 21, 2024 Published: January 22, 2025
Academic Editor: Andy Wai Kan Yeung, The University of Hong Kong, China
The article belongs to the special issue Digital Health Technologies for the Early Detection of Oral Cancer
Confocal laser endomicroscopy (CLE) enables real-time diagnosis of oral cancer and potentially malignant disorders by in vivo microscopic tissue examination. One impediment to the widespread clinical adoption of this technology is the need for operator expertise in image interpretation. Here we review the application of AI to automatic tissue classification of CLE images and discuss the opportunities for integrating this technology to advance the adoption of real-time digital pathology thus improving speed, precision and reproducibility.
Current management of oral potentially malignant disorders (OPMD) using clinical examination in conjunction with investigative biopsy and histopathology with long-term monitoring has remained essentially the same for many decades. The deficiencies in these approaches are well recognised and the way forward to improving patient outcomes in oral squamous cell carcinoma (OSCC) is earlier detection and better assessment of malignant transformation risk [1]. While biopsy and histopathological assessment are the gold standard, due to invasiveness such sampling must be used judiciously. Although the subjectivity of pathological assessment is well documented the clinicians’ selection of biopsy site is also a source of variability. Clinicians must make decisions about biopsy site(s) based upon conventional examination. Non-invasive microscopic imaging of tissues could significantly assist in surveying mucosal tissue for monitoring abnormality, reduce unnecessary biopsies, optimise site selection, and ultimately may supplant conventional biopsies. New technologies in non-invasive tissue imaging and computer-based image recognition are the key to driving these improvements. This applies not only to the application of artificial intelligence (AI) to enhance conventional histopathology techniques but also the introduction of clinical imaging technologies which synergise with AI for assessment of abnormality. Here, we discuss how AI image recognition can escalate the application of probe-based confocal laser endomicroscopy (CLE) to improve the diagnosis and management of OPMD, diagnosis of OSCC in the clinic, and determination of margins in surgery.
A number of in-clinic optical adjuncts have been shown to have efficacy in the clinical assessment and management of OPMD, including autofluorescence reflectance spectroscopy [2, 3] and wide-field narrow-band spectroscopy [4], but have not been widely adopted in clinical practice. These adjuncts provide clinicians with further guidance for assessment of oral mucosal lesions, however, they do not provide information on the cellular and architectural structures that are traditionally used to diagnose pathology [5]. One non-invasive tool that can survey tissue microarchitecture is CLE which combines endoscopic imaging with confocal microscopy [6]. In this fluorescence-based imaging method, excitation and image capture are achieved using a miniaturised probe which can be applied to the tissue surface to be imaged (Figure 1A). A fluorophore contrast dye (usually fluorescein) is typically applied topically prior to imaging in order to stain cellular structures such as nuclei and cell membranes. Commercially available hand-held CLE devices such as single-fibre distal scanning devices (Optiscan Imaging Ltd, Melbourne, Australia) enable real-time in vivo microscopic assessment of tissue at 1,000× magnification. An advantage of this method is that it provides microstructural assessment of tissues based upon similar criteria to those used in conventional histopathology (Figure 1B). Thus, interpretation of CLE images is based upon diagnostic principles already developed, refined, and widely accepted for oral epithelial dysplasia (OED) and OSCC.
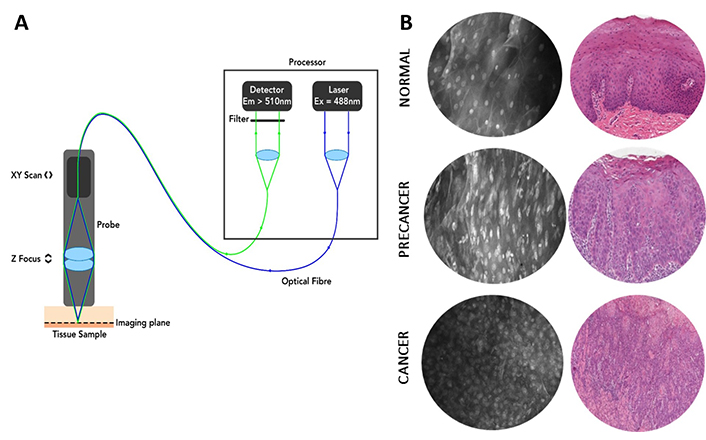
Principles and example images of probe-based CLE. (A) Diagram of the essential elements of single-fibre distal scanning CLE [confocal laser endomicroscopy (Optiscan Imaging Ltd, Melbourne Australia)]. Em: emission; Ex: excitation. Reprinted with permission from [7], © 2021 Microscopy Society of America. (B) Optical CLE sections obtained with ViewnVivo® by Optiscan and corresponding conventional hematoxylin and eosin histopathology show the ability of CLE to visualise cancer and precancerous tissue architecture. Reprinted with permission from Optiscan Imaging Ltd, internal instrument data, © 2025 Optiscan Imaging Ltd
In clinical practice CLE has been successfully applied in diagnostics in the intestine [8], oesophagus [9], colon [10], and neurosurgery [11]. Earlier pilot studies have demonstrated the feasibility of applying CLE imaging to oral lesions including oral leukoplakia and OSCC (reviewed in [12]). More recently, intraoperative CLE with an intravenous fluorescein contrast agent has been used to develop a scoring system based upon cellular morphology for classification of malignant tissue in the oral cavity [13]. In a recent clinical study, we have shown that in the oral cavity, CLE is a highly accurate (88.9%), easy-to-use, and rapid point-of-care technology for assisting clinicians in the diagnosis of OED/OSCC and discriminating between dysplastic and non-dysplastic pathology when measured against gold-standard histopathology using accepted World Health Organisation (WHO) architectural and cytological features of OED [14]. These specific features including nuclear pleomorphism, cellular pleomorphism, cellular crowding, and increased nuclear cytoplasmic ratio were used to make the CLE assessment. CLE imaging can also inform the presence of inflammatory infiltrate, keratinization, ulceration, or presence of micro-organisms such as fungal hyphae [14].
Despite the clear benefits of this technology in a number of settings, there are some impediments to its wider adoption. Interpretation of CLE images requires specific expertise which must be acquired through training and supervised experience just like histopathological interpretation of hematoxylin and eosin stained tissue samples. There is evidence that with appropriate training, surgeons and clinicians can reach a high level of competency in interpretation of CLE images [15, 16]. CLE generates digital images in a continuous fashion and therefore the ability of the operator to interpret a large number of images is crucial to its utility. This is an advantage in terms of diagnosis or detection of malignancy since CLE permits the operator to survey a large area of tissue or surgical margin but is limited by the operator’s capabilities to assess all images. In addition, because of the acquisition of images via a hand-held probe, this can lead to motion artefacts which can hamper diagnostic interpretation. One solution to these issues is telepathology where pathologists can interpret CLE findings for clinicians/surgeons by live-streaming CLE datasets [11]. While such an approach will enhance the uptake of CLE technology it still requires the availability of specialist expertise in image interpretation. It is for these reasons that the application of AI-based image recognition using machine learning (ML) algorithms for enhanced diagnostic imaging has been proposed and investigated to assist the non-specialist clinician/pathologist operator. Furthermore, such technology offers the opportunity to reduce subjectivity by standardisation of diagnostic features and facilitating interpretation. A potential added benefit is that AI-based image analysis may not only recognise established diagnostic features but may also identify visual characteristics not recognised by human visual analysis. In the clinical setting AI-assisted CLE imaging can assist not only in diagnosis but also in determining the extent of the abnormal tissue and be used to screen other anatomical locations where abnormality might be suspected upon examination.
The terminology around AI can be confusing since the terms AI, ML, and deep learning (DL) are often used interchangeably although it is best to consider ML as a subset of AI, and DL as a subset of ML. There are many excellent primers in this area [17–19]. The most common approach for application of AI to image analysis is by DL based upon various types of deep neural networks (NN) which consist of multiple layers of interconnected nodes trained by learning [17, 19]. NN can be applied to automatic classification and segmentation of cellular features. NN must be by definition functionally trained using data and most commonly in microscopy supervised learning is applied where NN are trained using labelled data. This requires a large number of diverse images which have been labelled and curated by human experts and is a significant barrier to the development of diagnostic NN models. If the training dataset is insufficiently large and diverse then the model may not be generalizable to real-world data [17]. Alternatively, in unsupervised learning the model identifies patterns and features using unlabelled data although a large dataset is still required, and this approach is less developed than supervised learning in pathology.
Because of the potential of AI to improve objectivity of current diagnostic approaches using visual examination in medical imaging and pathology, computer-aided diagnosis has been widely investigated in these fields [20, 21]. Recent reviews have examined the application of AI in the context of digital pathology [17], and specifically for the diagnosis of epithelial dysplasia [22]. The algorithmic approaches to visual analysis developed in these studies provide the basis for the development of AI tools for analysis of CLE images. The application of DL to CLE will be less complex in some respects than similar efforts in digital pathology. High-quality scanned whole-slide images (WSI) are too large and must be divided into smaller areas, a process known as image patch sampling [17]. This restriction does not apply to CLE images which are smaller, acquired continuously and can be used directly for training NN. One type of supervised NN that has been very successfully and broadly applied to medical image analysis and segmentation are convolutional NN (CNN) [17, 19], and this computational approach has been most commonly used for model development in CLE imaging.
In addition, computational tools developed for analysis of confocal microscopy in biomedical research where this technique is widely used also have relevance and applicability to clinical CLE diagnostics. For example, in one study the authors used sequential application of multiple NN to process raw images, to first denoise and then reduce artefacts by deconvolution and then perform segmentation to identify and analyse features [23]. Such AI-based systems developed to allow researchers to analyse large datasets of many complex confocal images indicate feasibility for clinical CLE image analysis.
A number of studies have reported application of supervised or semi-supervised ML methods to clinical CLE imaging of a variety of tissues. Many of these studies have used ex vivo tissue CLE imaging of resected specimens as proof of principle for in vivo applications although ex vivo examination does have utility for margin assessment in the surgical setting. In Barrett’s oesophagus where tissue surveillance is critical to clinical management, one recent study used DL models for diagnosis of dysplasia/cancer, finding performance consistent with reported human diagnostic accuracy [24]. In the gastrointestinal tract, where CLE-based methods are well established, promising results have been achieved with CLE images from animal models of colon cancer [25] and ex vivo gastric cancer tissue [26] using computational NN to identify malignant tissue. In the latter study, real-time evaluation using an AI-based method distinguished not only malignant from non-malignant tissue but also the histological subtype in gastric cancer [26]. In colon cancer and metastases, a CNN gave similar diagnostic performance in interpretation of CLE images to trained clinical observers suggesting that this approach could assist in intraoperative implementation of CLE [15]. In neurosurgery, AI-based classification has also been applied to identify glioblastoma or metastases in resected brain tumour tissue specimens, building a model with high accuracy (94%) [27]. Other neurosurgical studies using NN have similarly shown accurate classification of features in brain tissue CLE imaging (90% and 98.5%, respectively) [28, 29].
For OSCC there have been a number of reports of the development of ML tools to automate and improve consistency for interpretation of CLE images in the diagnostic [30] and surgical [31] settings. Aubreville et al. [30] successfully applied a CNN to in vivo CLE images of oral epithelium for identification of OSCC, reporting 88.3% accuracy. In the surgical setting, ML has been applied to ex vivo confocal imaging of resected OSCC as a means for in theatre assessment of surgical margins [31]. Large language models such as GPT have been explored in one pilot study of 5 patients for classification of CLE images in the diagnosis of oropharyngeal squamous cell carcinoma with a reported accuracy of 71.2% [32]. In a related field of high-resolution microendoscopy (HRME) with similar fluorescence images to CLE, a CNN-based approach has been used to quantify nuclear features and diagnose malignancy in oral epithelial images [33].
Apart from the histological classification of tissues, computational methods have been applied to artefactual features which can commonly impact the diagnostic utility of CLE images. To date, only a few studies have reported on the processing of CLE images although in the broader field of confocal microscopy, the literature is extensive. The automatic assessment and filtering of image quality using a DL model allowed the processing of a large dataset of oral cavity and vocal fold CLE images to distinguish those that were non-diagnostic and improve subsequent diagnostic accuracy to 94.8% [34]. Recently an approach based on a self-supervised algorithm incorporating multiple NN has been used to denoise CLE images and enhance image quality [35]. This essentially unsupervised method has advantages over supervised learning since it avoids the need to curate a large set of labelled training data. Examples of the applications of NN to CLE are illustrated in Figure 2.
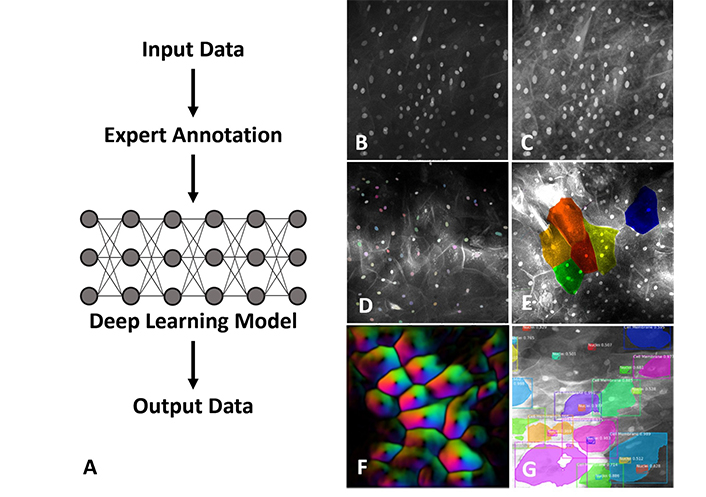
Examples of NN (neural network) application to imaging using single-fibre distal scanning CLE [confocal laser endomicroscopy (Optiscan Imaging Ltd, Melbourne Australia)]. (A) In supervised DL (deep learning) the model training process requires a large data set of expert-labelled images which can be developed for different image analysis tasks. An original captured image (B) can be enhanced and denoised by image pre-processing, filtering and masking for preparation of input (C). Morphological features and image features such as nuclei (D), cell boundaries (E) and optical vector intensity (F) can be labelled and mapped. Algorithms can use cell and nuclear size, pleomorphism, nuclear: cytoplasmic ratio, cell crowding, presence of inflammatory cells, and other features (G) to provide a diagnostic readout. Figures 2B–G are adapted with permission from Optiscan Imaging Ltd, internal instrument data, © 2025 Optiscan Imaging Ltd
These results show that computational automatic image analysis can yield results consistent with expert diagnosis and is likely to accelerate the application of CLE technology in OPMD and oral malignancy diagnosis and management. However, these investigations are generally pilot studies, and for wider acceptance of the integration of AI-based diagnostics and CLE imaging more extensive validation studies are required incorporating larger data sets with greater diversity than those reported to date.
A key determinant in the application of AI diagnostics in CLE is the quality of output images which may vary significantly between CLE technology platforms (single-fibre vs bundled-fibre). Most of the studies described thus far in the development of AI diagnostic models for application to CLE have used bundled-fibre-based systems which have some practical advantages but are disadvantaged by limited image resolution [7, 36]. Developing AI-based models for diagnosis of OED using such images has required the development of scoring systemswhich may not recapitulate commonly accepted cellular features of OED, such as those of the WHO. In contrast, single-fibre distal-scanning CLE has a number of technical advantages including superior image resolution and improved visualisation of cellular and nuclear features [7, 36]. Such CLE imaging platforms with high-quality image outputs are likely to be more amenable to AI diagnostics and require less image preprocessing. In addition, higher image quality greatly facilitates the training of ML models since expert labelling of training datasets is more straightforward. Furthermore, such AI diagnostic models are likely to be more acceptable to regulators and adopted by clinicians because they are readily explainable due to their alignment with existing diagnostic practice.
For CLE, AI is a transformative technology since it transforms CLE from an image acquisition device which requires expert interpretation to a true diagnostic device capable of providing a real-time point-of-care diagnostic readout during tissue examination. AI-based diagnostics of CLE images requires multiple NN integrated to perform sequential tasks to denoise/enhance images, label cellular features and generate diagnostic output. Computationally, the greatest requirements are in the development phase, where models must be trained using large, curated datasets. However, at the clinical application phase, sufficient computational resources must be integrated into the CLE platform so that the clinician or surgeon receives an immediate indication of abnormality as the probe is moved around the tissue surface. Prospectively, the availability of real-time digital pathology based upon CLE will have a significant impact on diagnostic and surgical workflows. Clearly, future studies will be needed to quantify the benefits of such workflows on patient outcomes and health care costs to provide objective evidence for adoption of this technology. To date there have been few studies of the health economic and workflow impacts of CLE imaging, however, those to date indicate that CLE in some settings can improve diagnostic accuracy, reducing costs and suggest that one of the principal barriers to the adoption of this technology which is the need for specific diagnostic expertise is one that can be addressed using AI [37, 38].
Looking to the future, one aspect of CLE imaging which has received some investigation and which could be integrated into AI-based workflow is the use of fluorescently labelled molecules to target and visualise specific biomarkers in vivo [39, 40]. Many studies have reported the application of AI assistance to bring greater objectivity and sensitivity to immunohistochemistry-based diagnostic applications (for example [41, 42]). Application of specific probes for biomarkers has been explored in CLE and in at least one study of direct relevance to OSCC, AI has been successfully applied to CLE image segmentation in oesophageal adenocarcinoma labelled with fluorescently labelled epidermal growth factor receptor binding peptides [43].
Taken further, development of AI models of disease for OED and OSCC with or without fluorescently labelled molecules could be pursued in animal models such as the 4-Nitroquinoline-1-oxide (4NQO) mouse model or the hamster cheek pouch carcinogenesis model utilising CLE images obtained from longitudinal studies of oral carcinogenesis in these animal models. Not only is this approach valuable for developing AI useable data, but it also assists in understanding the natural history of oral carcinogenesis taking a systems biology approach not otherwise possible in cross-sectional studies requiring animal sacrifice at interval timepoints and examination of tissues using traditional approaches on glass slides.
AI is already broadly demonstrating that it will revolutionise pathology workflows and improve reproducibility. The studies to date have shown the capability of AI in the diagnostic interpretation and classification of CLE images. Integration of AI into the CLE workflow can enable the adoption of real-time digital pathology directly in the clinic to empower clinical and surgical decision-making. The integration of AI into CLE is in its infancy and the size and scope of studies to date have been small. Technical developments and acceptance of clinical AI in related pathology and medical image analysis fields will help to drive application to CLE. The keys to advancing application to CLE will be the development of reliable and platform-specific AI tools and the demonstration of their diagnostic performance in large robust clinical studies.
AI: artificial intelligence
CLE: confocal laser endomicroscopy
CNN: convolutional neural network
DL: deep learning
ML: machine learning
NN: neural network
OED: oral epithelial dysplasia
OPMD: oral potentially malignant disorders
OSCC: oral squamous cell carcinoma
WHO: World Health Organisation
SAF and CSF: Conceptualization, Investigation, Writing—original draft, Writing—review & editing. Both authors read and approved the submitted version.
CSF is the Chief Executive Officer and Managing Director of Optiscan Imaging Ltd (ASX: OIL) and holds equity in the company. SAF is an employee of Optiscan Imaging Ltd.
The figures in this manuscript use images obtained with ethical approval by the Bellberry Human Research Ethics Committee (2023-03-359-PRE-1) following scientific and ethical review.
Informed consent to participate in the study was obtained from all participants.
Not applicable.
Not applicable.
Not applicable.
© The Author(s) 2025.
Open Exploration maintains a neutral stance on jurisdictional claims in published institutional affiliations and maps. All opinions expressed in this article are the personal views of the author(s) and do not represent the stance of the editorial team or the publisher.
Copyright: © The Author(s) 2025. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Xin-Jia Cai
Eduardo D. Piemonte ... Jerónimo P. Lazos
Kehinde Kazeem Kanmodi ... Jacob Njideka Nwafor
Romina Andrian ... Gerardo Gilligan
Caique Mariano Pedroso ... Alan Roger Santos-Silva
Márcio Diniz-Freitas ... Pedro Diz-Dios
Gerardo Gilligan ... Eduardo Piemonte
Constanza B. Morales-Gómez ... Víctor Beltrán
Anna Luíza Damaceno Araújo ... Alan Roger Santos-Silva
