Affiliation:
1Flinders Health and Medical Research Institute, Flinders University, Bedford Park 5042, Australia
ORCID: https://orcid.org/0000-0001-5421-6817
Affiliation:
1Flinders Health and Medical Research Institute, Flinders University, Bedford Park 5042, Australia
2Eye and Vision Health, Flinders University, Bedford Park 5042, Australia
ORCID: https://orcid.org/0009-0002-1340-8542
Affiliation:
1Flinders Health and Medical Research Institute, Flinders University, Bedford Park 5042, Australia
ORCID: https://orcid.org/0000-0003-2492-135X
Affiliation:
1Flinders Health and Medical Research Institute, Flinders University, Bedford Park 5042, Australia
ORCID: https://orcid.org/0009-0001-2890-5210
Affiliation:
1Flinders Health and Medical Research Institute, Flinders University, Bedford Park 5042, Australia
3SA Pathology, Flinders Medical Centre, Bedford Park 5042, Australia
ORCID: https://orcid.org/0000-0003-3276-9685
Affiliation:
1Flinders Health and Medical Research Institute, Flinders University, Bedford Park 5042, Australia
Email: jill.carr@flinders.edu.au
ORCID: https://orcid.org/0000-0002-1080-1472
Explor Immunol. 2024;4:376–393 DOl: https://doi.org/10.37349/ei.2024.00147
Received: December 13, 2023 Accepted: May 20, 2024 Published: June 24, 2024
Academic Editor: Roberta Bulla, University of Trieste, Italy
Aim: Severe dengue is correlated with a decrease in the circulating complement regulator, factor H (FH) and prior work has shown that dengue virus (DENV) infection induces FH mRNA but not FH protein release. Here, the mechanisms of this phenomenon were defined.
Methods: HEK293 cells were infected with DENV-2 and changes in FH mRNA and protein were analyzed by real time reverse transcription-polymerase chain reaction (RT-PCR), enzyme-linked immunosorbent assay (ELISA), and fluorescent microscopy. Additionally, cells were stimulated with size fractionated supernatants from DENV-infected cells, supernatant containing DENV non-structural protein-1 (NS1) without virus particles, and infections performed with or without the toll-like receptor-4 (TLR4) antagonist, TAK-242. Mass spectrometry was used to define the protein content of the fractionated supernatant, and treatment of cells with sialidase or heparinase was used to define cell-associated FH protein.
Results: DENV-infection induced full-length FH mRNA and cell-associated FH protein. Microscopy demonstrated membrane and intracellular-associated FH with a cytoskeletal and perinuclear localisation, in both DENV positive and uninfected neighboring bystander cells. Fractionation of cultured supernatant from DENV-infected cells demonstrated that secreted factors > 50 (kilodaltons) kDa induced FH mRNA and this could be blocked with TAK-242 but was not simulated by the TLR4 agonist, DENV NS1. Mass spectrometry detected DENV envelope, membrane and NS1, complement component 5 (C5), and complement FB, and indicated a > 20-fold increase in C4, inter-alpha-trypsin inhibitor heavy chain H2 (ITIH2), and alpha-2-macroglobulin in the > 50 kDa fraction from DENV-infected compared with conditioned media from uninfected cells. Sialic acid levels were unchanged and cleavage did not affect release of FH from DENV-infected compared to uninfected cells. In contrast, sulphated glycosaminoglycans (GAGs) were reduced in the cultured supernatant and cell lysates following DENV-infection, and heparinase cleavage released significantly more FH from DENV-compared with uninfected cells.
Conclusions: Following DENV-infection, secreted molecules induce FH that remains intracellular and with increased binding to cell surface heparan sulphate. The mediators of induction of FH mRNA act in trans and via TLR4 but this is not likely to be via DENV NS1. The retention of FH in the local environment of the infected cell could benefit the virus by negating local complement killing of cells, and/or benefit the host by inhibition of heparan sulphate-mediated DENV infection to restrict viral spread.
Dengue virus (DENV) infection causes dengue disease, and is a significant and growing health burden, particularly in tropical and subtropical climates where the mosquito vector is present [1, 2]. Dengue can present with symptoms that range in severity from subclinical to life threatening, with the hallmarks of dengue being fever, vascular leakage, and decreased platelets. Disease can be classified as dengue, dengue with warning signs, and severe dengue [3]. Numerous factors influence disease severity, including immune activation and excessive cytokine production [4, 5]. Actions of the DENV protein, non-structural protein-1 (NS1) are likely important, particularly as an inciting trigger for disruption of the endothelial cell (EC) barrier of the vasculature that can lead to vascular leak syndrome [6–8]. Additionally, the complement system can contribute to immune dysregulation, where increased dengue disease severity has been associated with increased levels of serum complement proteins, factor D, and a decrease in the negative regulator of the complement alternative pathway (AP), factor H (FH) [9, 10]. The role of the complement system in viral infections such as dengue has been reviewed [11, 12].
The complement system defines three pathways that are activated via a series of proteolytic cleavages: the classical (CP), mannose-binding-lectin (MBL), and the AP. Each pathway converges at complement component 3 (C3) cleavage to activate the common terminal pathway to form the membrane attack complex (MAC), which causes lysis of pathogens or infected cells [13]. Poorly regulated complement can result in immune-mediated pathologies through self-targeted MAC and excessive complement-derived anaphylatoxins produced by cleavage of C3 and C5, that act as potent chemoattractants with vasoactive properties. Complement activation positively feeds-back through the AP which accounts for a major portion of total complement activity [14]. Thus, controlling the AP is crucial in preventing complement-mediated immunopathology. FH is the most important negative regulator of the AP and acts by binding host-cell ligands, including sialic acid and sulphated glycosaminoglycans (GAGs), to positively regulate the cleavage of C3b and prevent further C3 cleavage by the AP C3 convertase and activation of the downstream terminal complement pathway. Reduced FH and hence ability to modulate AP activity can cause serious diseases such as nephropathy and coagulopathies [15, 16], and analysis of dengue patient serum has revealed significantly less serum FH in patients with severe dengue compared to dengue fever [9].
Our laboratory has defined increased FH mRNA following DENV infection without an accompanying increase in FH protein release into the cultured supernatant from primary macrophages and EC in vitro. Importantly, this was observed concomitantly with increased secretion of the AP activator, complement FB, and elevated ability to stimulate AP activity in vitro [10]. Although the liver is the main producer of circulating FH, monocyte-macrophages are the primary target for DENV-infection and EC are the target of pathology in vivo. Thus, understanding how local FH production from these cell types and AP regulation is influenced by DENV infection is important [17, 18]. To investigate this further, the study here has validated HeLa cells as a model where DENV-infection induces FH mRNA but not FH protein released into cultured media, as seen in primary DENV-target cells. Using this model, it was determined that DENV infection induces a secreted factor, > 50 (kilodaltons) kDa, that induces FH mRNA via toll-like receptor-4 (TLR4). FH protein, however, is retained intracellularly along the cytoskeleton and around the nucleus, and extracellularly bound to heparan sulphate on the cell surface in both infected and uninfected bystander cells. These results suggest that in the local environment of a DENV-infected cell, FH is induced but not released into the extracellular milieu and perhaps serves to protect these cells from complement-mediated killing or to restrict viral spread.
HEK293, human embryonic kidney cells, HeLa, uterine epithelial cells and Huh7.5, liver cells (kindly supplied by Professor Michael Beard, The University of Adelaide) were cultured in DMEM (Gibco, ref # 11965-092) or U937 promonocytic cells in RPMI-1640 (Gibco, ref # 11875-093) supplemented with 10% (v/v) fetal calf serum (FCS), 100 units penicillin, 0.1 mg streptomycin and 2 mM GlutaMAX (Thermofisher). Cell lines were from American Type Culture Collection (ATCC). Cells were passaged every three and four days and validated to be Mycoplasma spp. negative by PCR. Short tandem repeat (STR) profiling was undertaken for HeLa cells (Australian Genomics Research Facility, AGRF, Melbourne, Victoria), with a worst-best match of 87.5–100%, authenticating this cell line. HEK293 cells containing a DENV subgenomic green florescent protein (GFP) reporter genome were generated and termed HEK-pREP, as described previously [19] from constructs provided by Professor Ted Pierson [20].
DENV infections were performed with the laboratory DENV-2 clone, Mon601, derived from the New Guinea C strain [21]. DENV stocks were generated in C6/36 insect cells, and titres were quantitated by plaque assay, as described previously [19]. Cells were seeded the day prior to infection with DENV at a multiplicity of infection (MOI) of 1 for 90 minutes (min) at 37°C. Following infection, inoculum was removed, and cells were washed and cultured in complete media at 37°C with 5% CO2. Supernatant, fixed cells, or RNA was harvested at indicated time points and analysed as below. For experiments to assess the role of TLR4, cells were incubated from 2 hours (h) prior to infection up to 24 h post-infection [hours post infection (hpi)] with 1 µM TAK-242 (Selleck Chemicals). At 24 hpi RNA was extracted for analysis of FH by reverse transcription-quantitative polymerase chain reaction (RT-qPCR), as below.
Total RNA was extracted from cells lysed in TRIsure reagent (Bioline), DNAse I (Zymo Research) treated, and 500 ng was reverse transcribed with Moloney murine leukemia virus RT and random hexamers (NEB). The cDNA was diluted and assayed by PCR using iTaq (BioRad) with primers as shown in Table 1. qPCR was performed on a Rotor-gene iQ (Qiagen) with cycling parameters: 1 cycle of 95°C for 5 min followed by 40 cycles of 95°C for 15 s, 58°C for 20 s, 72°C for 20 s followed by a melt analysis to validate the product. Results were normalised to the housekeeping gene, cyclophilin, and relative changes in mRNA abundance were calculated using the double delta cycle threshold (Ct) method and normalized against uninfected or untreated controls [22]. PCR controls included an FH clone constructed previously [23] and water as a no template negative control.
Primer sequences were used for reverse transcription-polymerase chain reaction (RT-PCR) and to discriminate human factor H (FH), FH-like-1 (FHL-1), and FH-related protein-1 (FHR-1)
| Primer name | Sequence | mRNA target |
|---|---|---|
| FH short consensus repeat 2 (SCR2) | Forward: AGGCCCTGTGGACATCReverse: AACTTCACATATAGGAATATC | FH and FHL-1, across exons 3–4, SCR2 (5’ end) |
| FH exon 9/11 | Forward: CTACTCCCAGATGCATCCGTGTReverse: GGTTGAGCTGACCATCCATCTTT | FH, across exons 9–11 (middle) |
| FH SCR20 | Forward: CATCCGTGTGTAATATCCCGReverse: TCTTTTTGCACAAGTTGGATAC | FH, across exons 22–23, SCR20 (3’ end); potentially FHR-1 across exons 5–6 |
| FHL-1 | Forward: CATCCGTGTCAGCTTTACCCTReverse: CCATTCTGGCTGGAATAATACAC | FHL-1 unique 4 amino acid tail |
| FHR-1 | Forward: GTACTGTGGGCGGATTCACAReverse: CTCTGTTGGGGGAGAAGAAAAATA | FHR-1 A and B, exons 1–4 |
| Cyclophilin | Forward: GGCAAATGCTGGACCCAACACAAAReverse: CTAGGCATGGGAGGGAACAAGGAA | Cyclophilin A, housekeeping gene |
FH enzyme-linked immunosorbent assay (ELISA) was performed as previously [10]. In brief, assays utilised goat anti-FH capture antibody (Calbiochem, Cat# 341276) and detection of complexes with mouse anti-FH (Abcam, Cat# ab17928) followed by goat anti-mouse IgG-horse radish peroxidase (HRP) conjugate (Santa Cruz, Cat# sc-17951) and colourimetric detection at 450 nm (Spectramax® ID5, Molecular Devices). FH concentration was determined from a standard curve of purified FH (10 μg/mL to 10 ng/mL).
Dimethyl methylene blue (DMMB, Sigma) reagent was prepared as described previously [24]. In brief, DMMB (38 μM), glycine (40.5 mM), and NaCl (27.4 mM) were dissolved in 0.01 M acetic acid and filtered through a 0.45 μM filter (Sartorius). Immediately prior to use, 2 M Tris pH 7.2 was added to DMMB reagent (1:4), then reacted with 5 μL of undiluted supernatant or cell lysate. Absorbance at 525 nm was measured immediately (Spectramax ID5, Molecular Devices), and values were determined by comparison to a standard curve of chondroitin sulphate (Sigma, Cat# C9819) from 30μg/mL to 500 μg/mL.
Cells were cultured and infected with DENV or left uninfected as described above. At 48 hpi supernatants were removed and replaced with phosphate buffered saline (PBS) containing 5 mM CaCl2 with or without 400 μIU/mL Heparinase I (Sigma, Cat# H2519) and 0.415 mIU/mL Heparinase III (Sigma, Cat# H8891) and incubated for 4 h at 37°C in 5% CO2. Supernatants were harvested for the quantitation of FH by ELISA.
Cells were seeded onto a 96-well opaque-walled plate (Perkin Elmer, Cat# 6055300) at a density of 3 × 104 cells/well in complete DMEM and incubated overnight at 37°C 5% CO2. Cells were then either uninfected or DENV-infected (MOI = 1) as above. At 48 hpi, cells were fixed using 2% (w/v) paraformaldehyde (PFA), permeabilised for 20 min with 0.05% (v/v) IGEPAL (Sigma, Cat# 542334), and concomitantly stained with Hoechst 33342 (5 μg/mL) and FITC-conjugated Sambucus nigra sialic acid binding lectin (25 μg/mL, Thermofisher, Cat# L32479). Fluorescence was measured using a spectrophotometric plate reader (SpectraMax iD5) and fluorescence for sialic acid (495ex/519em) was normalised to fluorescence of nuclei at (361ex/497em).
Cells were uninfected or DENV-infected and at 48 hpi fixed with PFA 2% (w/v) for 10 min at 4°C, rinsed with 70% ethanol (v/v), and stored in PBS. For immunostaining, cells were blocked with 1% (w/v) bovine serum albumin (BSA) and 2% (v/v) normal donkey sera diluted in PBS and incubated at room temperature for 30 min. Cells were permeabilised in 0.5% (v/v) IGEPAL for 15 min at room temperature, then co-stained with polyclonal goat anti-FH (Calbiochem) and mouse anti-dsRNA (English and Scientific Consulting) at 1/100 and 1/200 dilution, respectively overnight in a humidified chamber. Cells were washed with PBS and labelled with secondary antibodies (rabbit anti-goat Alexa-555 and rabbit anti-mouse Alexa-488, Thermofisher) and Hoechst 33342 (5 μg/mL). Slides were mounted with Prolong gold anti-fade (Invitrogen) and viewed using either an AX70 fluorescence microscope (Olympus) or LSM880 Confocal Microscope (Zeiss). Images were captured using Zen Blue 3.0 software (Zeiss) and rendered in Imaris 10.0 (Oxford Instruments) which was also used to generate MP4 digital multimedia images. For detection of NS1, HEK293 or HEK-pREP were fixed and immunostained, as above, with 4G4 anti-DENV NS1 monoclonal antibody at 1/200 dilution (Uniquest).
HeLa cells were seeded at a density of 1 × 106 cells in a 75 cm3 flask for 24 h prior to DENV-infection as above at (MOI = 1). Inoculum was then removed, and cells were incubated for 48 h in complete media. Following incubation, supernatants were harvested, cellular debris removed by centrifugation, and then fractionated into > 50 kDa proteins using Ultra-15 Centrifugal filters (Amicon), for 15 mins at room temperature, at 2,600 × g. Fractions were reconstituted to the starting volume in complete media and stored at –80°C until required. For generation of NS1-containing supernatant, media was collected from a confluent flask of HEK-293 cells or HEK293-pREP. Harvested supernatant was clarified by centrifugation and stored at –80°C. Supernatant was subjected to non-reducing sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and western blot, with 4G4 anti-DENV NS1 at 1/10 dilution (Uniquest). Bound complexes were detected by chemiluminesence (BioRad, Clarity) and imaged (ChemiDoc Touch Imaging system, BioRad). For stimulation of cells with fractionated supernatant or NS1, HeLa cells were seeded the day prior to experimentation at a density of 3 × 105 cells per well in a 6-well plate (Nunc) and then stimulated by addition of undiluted supernatant generated above. At 8 h post-stimulation, cells were harvested in TRIsure and stored at –80°C for total RNA extraction and RT-qPCR, as above.
For mass spectrometry analysis, cultured supernatant was harvested and fractionated, as above but in the absence of FCS. Total protein within the supernatant was quantitated (Bio-Rad Protein Assay) and 10 μg of protein was processed with Sera-Mag Carboxylate SpeedBeads, (Cytiva) and digested overnight at 37°C using 0.5 μg/mL trypsin gold (Promega Corporation, Alexandria, NSW, Australia). Digested proteins were prepared for mass spectrometry by clean up again using Sera-Mag Carboxylate SpeedBeads, peptides dissociated and analysed by the Flinders Proteomics Facility, using Mass spectrometry (TSQ AltisTM Triple Quadrupole Mass Spectrometer, Thermo Scientific) as previously described [25]. Protein species were identified using Proteome Discoverer software (ThermoScientific). Raw abundance was normalised to total protein and summary peptide scores were recorded. Proteins with a peptide summary score < 5 were removed from consideration. Raw data available at: https://zenodo.org/record/6911339#.YuDXjHZByUl.
Data was analysed for normal Gaussian distribution using the D’Agostino and Pearson or Anderson-Darling test. Normally distributed data was analysed by one-way or two-way analysis of variance (ANOVA) followed by Bonferroni correction or Student’s t-test. Non-normally distributed data were analysed by Kruskal-Wallis test with Dunn Multiple comparsion or Mann-Whitney u-test. Data were using Prism 10.0.0 (GraphPad). All graphs are representative of average values ± standard error of the mean (SEM).
Experiments in primary macrophages, as previously undertaken [10], are difficult, and thus a cell line was sought that reflected the same responses of FH to DENV infection, as seen in macrophages. Analysis of uninfected or DENV-infected U937 and HEK293 cells did not detect FH mRNA or protein (data not shown). In contrast to our prior findings in primary macrophages, DENV-infection of the liver cell line Huh7.5 resulted in a significant decrease in FH mRNA that was accompanied by a significant reduction in FH protein in the supernatant (Figure 1A) and in cells as detected by immunofluorescent staining and microscopy (Figure 1B). As seen in DENV-infected primary macrophages, DENV-infection of HeLa induced increased FH mRNA, but unchanged secreted FH compared to uninfected controls (Figure 2A). When assessed by immunofluorescent staining and microscopy, however, increased FH protein was evident that was associated with cells (Figure 2B).
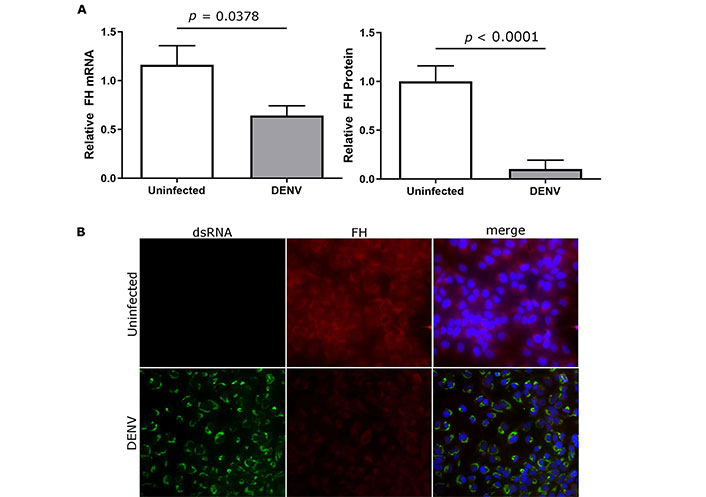
DENV-infection reduces FH mRNA, supernatant, and cell-associated FH protein in Huh7.5 cells. Huh7.5 cells were uninfected or DENV-infected and at 48 hpi (A) total cellular RNA, or supernatant were collected and analysed for FH mRNA by RT-qPCR (left panel) and supernatant FH protein by ELISA (right panel). qPCR results were normalised to cyclophilin, and protein and mRNA levels were expressed relative to uninfected cells. Results represent the mean ± SEM for triplicate samples from three independent infections and analysed by Student’s t-test; (B) stained for nuclei, double stranded RNA, and FH. Representative images are presented and captured at 40× magnification. DENV: dengue virus; FH: factor H
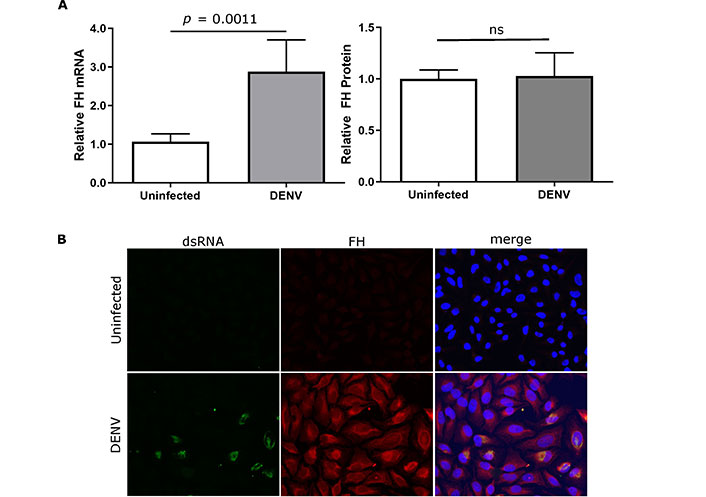
DENV-infection induces FH mRNA and cell-associated but not supernatant-associated FH in HeLa. HeLa cells were uninfected or DENV-infected and at 48 hpi (A) total cellular RNA, or supernatant were collected and analysed for FH mRNA by RT-qPCR (left panel) and supernatant FH protein by ELISA (right panel). qPCR results were normalised to cyclophilin, and protein and mRNA levels were expressed relative to uninfected cells. Results represent the mean ± SEM for triplicate samples from three independent infections and analysed by Mann-Whitney u-test; (B) stained for nuclei, double stranded RNA, and FH. Representative images are presented and captured at 40× magnification. ns: no significant changes observed. DENV: dengue virus; FH: factor H
Based on these results, FH responses to DENV in HeLa cells are reflective of those in primary macrophages, and HeLa cells were used for further study. Since the 4 kilobase (kb) FH mRNA can be alternatively spliced to produce a smaller FH-like-1 (FHL-1, 1.4 kb) mRNA that is identical to FH mRNA between short consensus repeat 1 (SCR1) and SCR7 but contains a unique 4 amino acid terminus (Figure 3A), studies characterised the forms of FH/FHL-1 mRNA produced. Additionally, there is high homology between FH SCR20 and SCR5 of FH related proteins FHR-1A and FHR-1B (Figure 3A) and thus primers were designed to detect the unique centre of FH transcripts (exons 9/11), the 3’ terminus found in both FH and FH-related protein-1 (FHR-1, FH SCR20) and the unique 3’ region of FHL-1 transcripts (Figure 3A). Results from all the three FH primer sets demonstrated a significant increase in FH mRNA in DENV compared to uninfected HeLa cells, with a comparable fold increase measured for each PCR (Figure 3B). Additionally, FHL-1 and FHR-1 mRNA were not detected in uninfected or DENV-infected HeLa cells. Thus, mRNA changes detected by PCR reflect increases in full-length FH mRNA and not FHL-1, or FHR-1 mRNA, and the immunoreactive protein detected by staining and microscopy (Figure 2B), likely represents FH and not cross reactive FHL-1 or FHR-1 proteins.
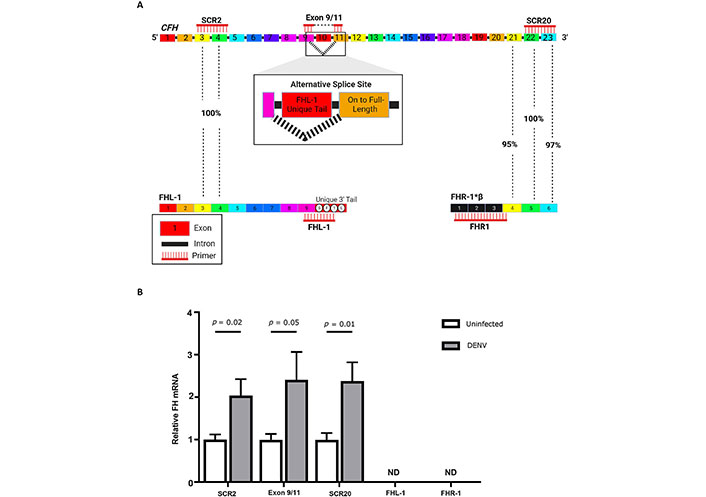
Full length FH mRNA is increased in DENV-infected cells and mRNA’s for FHL-1 and FHR-1 are not detectable. (A) The CFH gene produces FH mRNA which will be detected by primers for SCR2, exons 9/11, and SCR20. The FH transcript can be alternatively spliced to produce FHL-1, which will be detected by primers for SCR2 and uniquely by FHL-1 primers targeted at the 3’ tail. Both isoforms FHR-1A and FHR-1B mRNA can be detected by SCR20 primers due to high sequence identity between FH SCR20 with FHR-1, SCR5, and SCR6. FHR-1A and FHR-1B are uniquely targeted by primers for FHR-1 exons 1–4; (B) mRNA was quantitated in uninfected and DENV-infected HeLa by RT-qPCR using primers described in A. Results were normalised to cyclophilin and expressed relative to uninfected cells. Results represent the mean ± SEM for triplicate samples, from three independent infections, and analysed by Student’s t-test. CFH: complement factor H; DENV: dengue virus; FH: factor H; FHL-1: FH-like-1; FHR-1: FH-related protein-1; ND: Not detected; SCR2: short consensus repeat 2
Cell-associated FH protein was next quantitated by ELISA and results demonstrate increased FH protein in lysates from DENV-infected compared to uninfected cells (Figure 4A). FH immunostaining and confocal microscopy demonstrated low intensity fluorescence mainly localised to the cell surface in uninfected HeLa cells (Figure 4B and 4C). In contrast, FH staining in the cells from the DENV-infected cultures demonstrated increased fluorescence intensity at the cell surface, in the perinuclear zone around the nucleus, and intracellularly, along cell networks (Figure 4B and 4C). This distribution was observed in both dsRNA positive and negative cells of the DENV-challenged culture, suggesting induction in trans and potentially via release of a secreted factor (Figure 4B and 4C). To investigate this, conditioned media from uninfected HeLa or supernatant from DENV-infected HeLa that had been fractionated into proteins > 50 kDa was added to naive HeLa cells. Direct DENV infection of HeLa was used as a comparative control. At 8 h post treatment cellular RNA was harvested and FH mRNA was quantitated. DENV-infected cells, or treatment with the unfractionated supernatant and < 50 kDa fraction from DENV-infected cells did not induce a significant increase in FH mRNA (Figure 4D). The > 50 kDa fraction from DENV-infected cells, however, induced significantly higher FH mRNA compared to conditioned media from uninfected cells (Figure 4D).
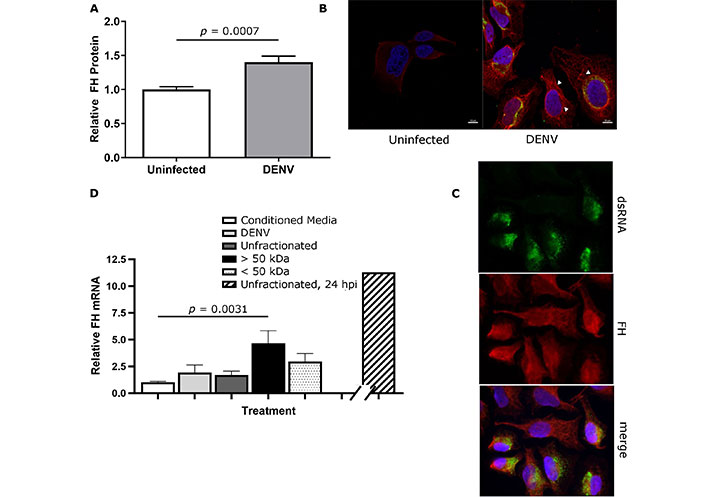
Cell-associated FH is increased in DENV-infected and uninfected bystander cells. HeLa cells were uninfected or DENV-infected and at 48 hpi, (A) total protein was extracted, and FH quantitated by ELISA, normalised against total protein and expressed relative to uninfected cells and analysed by Student’s t-test; (B) and (C) cells were fixed, and immunostained for FH and dsRNA. Nuclei were stained with Hoechst-33342. Representative images were captured with a Zeiss LSM880 confocal microscope at 60× magnification. FH association with the cell membrane, perinuclear and intracellular networks is indicated by arrowheads in both dsRNA negative bystander and dsRNA positive cells; (D) HeLa were treated with conditioned media, DENV infected (DENV), unfractionated, > 50 kDa or < 50 kDa fractionated supernatant from DENV-infected cells. RNA was harvested 8 h post-treatment for RT-qPCR for FH mRNA. Results were normalised to cyclophilin and expressed relative to conditioned media controls. Bar on broken x-axis represents the DENV and FH mRNA levels quantitated following stimulation with unfractionated supernatant at 24 h, as a comparator. Results represent the mean ± SEM from three independent experiments and analysed by Kruskal-Wallis test with Dunn multiple comparison. DENV: dengue virus; FH: factor H; hpi: hours post infection; kDa: kilodaltons
Next, mass spectrometry was performed to assess the secreted proteins present in the > 50 kDa supernatant fraction from DENV-infected cells. Excluding low confidence reads (SUM PEP score < 5), 232 secreted extracellular proteins were detected in the conditioned media from uninfected HeLa cells, and 102 secreted extracellular proteins were identified in the > 50 kDa supernatant fraction from DENV-infected cells. Although some < 50 kDa proteins were observed in the > 50 kDa fraction, these were minor. Of the 102 secreted and extracellular host-proteins, 9 were unique in the > 50 kDa supernatant fraction from DENV-infected cells and not detected in the conditioned media, including the complement proteins C5 and FB (Table 2). As expected, the > 50 kDa fraction of the DENV-infected HeLa supernatant contained DENV structural proteins: membrane and envelope, as well as NS1, which is a secreted DENV protein (Table 2). Although these results were from a single mass spectrometry analysis, proteins with > 20-fold-changes in > 50 kDa compared to conditioned media were detected. alpha2-Macroglobulin (alpha2M) was a highly abundant protein (2nd percentile) in the > 50 kDa fraction but far lower in the conditioned media (27th percentile). Similarly, inter-alpha-trypsin inhibitor heavy chain H2 (ITIH2, 3rd percentile) and C4 (34th percentile) were abundant in the > 50 kDa fraction but either low or approaching the lower limit of detection in the conditioned media (47th and 88th percentiles respectively). No cytokines were detected by mass spectrometry in the > 50 kDa fraction of the DENV-infected supernatant or the uninfected conditioned media, and FH levels by mass spectrometry were comparable in the conditioned media from uninfected cells and the > 50 kDa fraction from DENV-infected cells, reinforcing the quantitation of FH in cultured supernatant by ELISA (Figure 1). To test if DENV NS1 protein was a mediator of FH mRNA induction, cells were treated with NS1 in supernatant derived from HEK293 harbouring a subgenomic DENV replicon, termed here HEK-pREP. The presence of NS1 in HEK-pREP but not parental HEK293 was validated by immunofluorescent staining (Figure 5A) and western blot (Figure 5B). HeLa cells were then left unstimulated or stimulated with supernatant from HEK-293 control or HEK-pREP and FH mRNA was quantitated. Results indicate that treatment with supernatant containing NS1 does not affect induction of FH mRNA (Figure 5C). Since NS1 reportedly acts via TLR4 to elicit pathogenic responses, cells were DENV infected, cultured in the presence of the TLR4 antagonist, TAK-242 and FH mRNA quantitated. Results demonstrate that DENV-induction of FH mRNA was blocked by the addition of TAK-242 (Figure 5D).
Proteins that were uniquely detected and at > 20-fold relative abundance in the > 50 kilodaltons (kDa) fractionated supernatant from dengue virus (DENV)-infected cells compared to conditioned media
| Category | Gene name | Ranked abundance > 50 kDa (percentile) | Peptide score > 50 kDa | Conditioned media ranked abundance (percentile) | Conditioned media peptide score | Molecular weight (kDa) | Protein identified | Abundance in > 50 kDa fraction | Abundance in conditioned media | Relative abundance |
|---|---|---|---|---|---|---|---|---|---|---|
| Cellular proteins | APOB | 26 (26%) | 31.91 | - | - | 489.5 | Mutant Apo B 100 | 7.34E + 07 | ND | - |
| B4GALT1 | 89 (89%) | 39.789 | - | - | 43.9 | Beta-1,4-galactosyltransferase 1 | 2.67E + 06 | ND | - | |
| C5 | 40 (40%) | 12.32 | - | - | 188.2 | Complement C5 | 3.71E + 07 | ND | - | |
| FB | 99 (99%) | 5.33 | - | - | 85.5 | Complement factor B | 1.02E + 06 | ND | - | |
| COL2A1 | 68 (68%) | 7.757 | - | - | 141.7 | Collagen alpha-1(II) chain | 7.81E + 06 | ND | - | |
| CPN1 | 83 (83%) | 5.762 | - | - | 52.3 | Carboxypeptidase N catalytic chain | 3.79E + 06 | ND | - | |
| *HBA2 | 14 (14%) | 26.337 | - | - | 15.2 | Mutant haemoglobin alpha 2 globin chain | 1.54E + 08 | ND | - | |
| MMP2 | 32 (32%) | 15.609 | - | - | 73.8 | Matrix metallopeptidase 2 | 5.39E + 07 | ND | - | |
| P4HB | 69 (69%) | 25.263 | - | - | 55.3 | Protein disulphide-isomerase | 7.61E + 06 | ND | - | |
| DENV proteins | DENV E | 54 (54%) | 3.634 | - | - | 54.2 | Envelope protein | 1.44E + 07 | ND | - |
| DENV M | 28 (28%) | 8 | - | - | 379.3 | Membrane Protein | 6.78E + 07 | ND | - | |
| NS1 | 17 (17%) | 31.45 | - | - | 379.5 | Non-structural protein 1 | 1.17E + 08 | ND | - | |
| > 20-fold increase in abundance | ITIH2 | 3 (3%) | 30.269 | 108 (47%) | 10.261 | 106.4 | Inter-alpha-trypsin inhibitor heavy chain H2 | 1.09E + 09 | 1.73E + 07 | 63.01 |
| C4A | 34 (34%) | 7.954 | 204 (88%) | 6.553 | 187.6 | Complement C4 | 5.04E + 07 | 1.35E + 06 | 37.32 | |
| Α2M | 2 (2%) | 44.999 | 62 (27%) | 97.315 | 163.2 | Alpha-2-macroglobulin | 1.49E + 09 | 4.36E + 07 | 34.11 | |
| Unchanged | FH | 50 (50%) | 19.189 | 60 (26%) | 78.531 | 139 | Complement factor H | 1.60E + 07 | 5.27E + 07 | 0.30 |
*: < 50 kDa protein, DENV proteins are indicated, FH is shown for comparison; -: no data. ND: Not detected
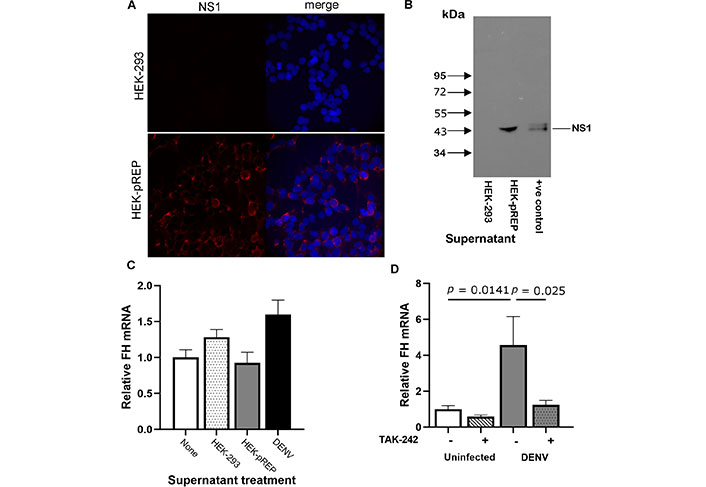
TAK-242 inhibits FH mRNA induction from DENV-infected HeLa but FH mRNA is not induced via NS1. (A) HEK293 or HEK cells containing a subgenomic replicon (HEK-pREP) were fixed and stained for NS1. Representative images were captured with an Olympus AX70 Fluorescence microscope at 20× magnification; (B) supernatant was collected from HEK293 or HEK-pREP and subjected to sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and western blot for NS1; +ve: positive control; (C) supernatant was used to treat HeLa cells or cells were DENV-infected (control). At 8 h post-treatment RNA was extracted and subjected to RTqPCR for FH mRNA. Results were normalized to cyclophilin, expressed relative to untreated cells, and represent mean ± SEM, n = 3, no significant changes observed; (D) HeLa were uninfected or DENV-infected in the presence (+) or absence (–) of 1 µM TAK-242. At 24 hpi, RNA was harvested and analysed for FH mRNA by qRT-PCR. Results were normalised to cyclophilin, expressed relative to FH mRNA levels in uninfected HeLa, and represent mean ± SEM from three independent experiments, analysed one-way ANOVA with Bonferroni correction. DENV: dengue virus; FH: factor H; kDa: kilodaltons; NS1: non-structural protein-1
In addition to secreted proteins, GAG and sialic acid—ligands that can both bind FH, were quantitated in response to DENV-infection. A fluorescein-conjugated Sambucus nigra sialic acid binding lectin-based assay was used to quantitate membrane-bound sialic acid, and sialic acids were cleaved by treatment with neuraminidase. No significant difference in sialic acids was detected between uninfected and DENV-infected cells (Figure 6A) or following treatment of cells with neuraminidase (Figure 6B). Quantitation of total sulphated GAGs demonstrated significantly less GAGs in the supernatant and lysates from DENV-infected compared to uninfected cells (Figure 6C). Cleavage of heparan sulphate from cells increased the amount of FH in the supernatant for both uninfected and DENV-infected cells. There was, however, significantly more FH released from DENV- compared to uninfected cells following heparinase treatment (Figure 6D).
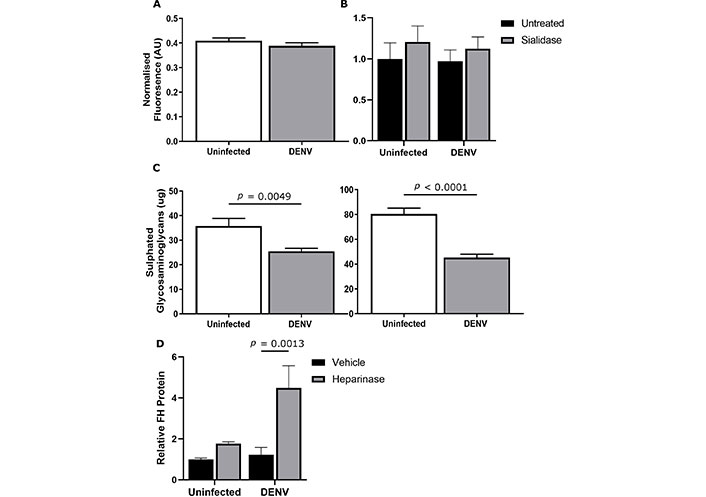
DENV infection does not impact sialic acids but reduces sulphated glycosaminoglycans (GAGs) which bind FH on DENV-infected cells. HeLa was uninfected or DENV-infected and at 48 hpi (A) cells were fixed, and cell surface sialic acid was detected with FITC-labelled Sambucus nigra binding lectin. Fluorescence [arbitrary units (AU)] was quantitated and normalised to nuclear staining. Results are representative of the means ± SEM of three separate experiments (n = 40), no significant changes were detected; (B) cells were untreated or sialidase treated (Clostridium perfringens neuraminidase, 90 mIU/mL) for 1 h and FH released into the supernatant quantitated by ELISA. Results were normalized to uninfected, untreated control, no significant changes were detected; (C) dimethyl methylene blue (DMMB) assay was used to quantify sulphated GAGs in the cell culture supernatant (left panel) and cell lysates (right panel). Results are representative of the mean ± SEM of three individual experiments, analysed by Student’s t-test; (D) cells were treated with PBS + 5mM CaCl2 (vehicle) or heparinase I and III (heparinase). FH released into the supernatant was quantitated by ELISA and normalised to uninfected, vehicle controls. Results represent the mean ± SEM from three independent, and analysed by two-way ANOVA with Bonferroni corrections. DENV: dengue virus; FH: factor H
Dengue disease severity has been previously correlated with increased complement activity and decreased circulatory FH [9]. Our laboratory has demonstrated that DENV infection in primary macrophages induces mRNA for both FB and FH but results in increased complement AP activity in cell supernatants due to an increase in secreted FB but not FH protein [10]. Here, our study has further defined this lack of FH protein release from DENV-infected cells. Since studies in primary macrophages are complex to undertake at scale and donor genetic and environmental variability can influence experimental responses, a stable cell line that reproduced the phenomenon of DENV-induction of FH mRNA but not released FH protein, was sought. The pro-monocytic cell lines, U937, and HEK293 produced no detectable FH mRNA or protein and were not considered further. Surprisingly and in contrast to primary macrophages, DENV-infection of the liver cell line Huh7.5, resulted in decreased FH mRNA and protein. The liver is the main producer of circulating FH. Although Huh7.5 cells are a hepatoma cell line, these cells are deficient in pathways that respond to type I interferon (IFN). IFN is a known regulator of FH [26] and computational analysis of the CFH promoter has identified a strong dependency on IFN-stimulated response elements [27, 28]. Thus, the difference in FH mRNA induction between Huh7.5 and macrophages may result from IFN-response deficiency of Huh7.5 or reflect hepatic-specific responses and true differences in FH regulation between DENV-infected macrophages and the liver [26]. The decreased FH production by Huh7.5, however, mimics the observed decreased circulating FH in severe dengue patients, and hence this is of interest to investigate further [9]. With regards to the biology of FH and DENV in macrophages, an important site for DENV replication in vivo, HeLa reflected the same phenomena, with increased FH mRNA but unchanged FH secretion into the supernatant. Thus, HeLa cells were selected as a model to focus on the molecular mechanisms of FH responses to DENV infection in macrophages.
To test if the increase in FH mRNA but lack of increase in secreted FH protein was due to a lack of full-length FH mRNA, including induction of known alternative splicing transcripts to generate FHL-1 mRNA or potential detection of homologous regions of the FHR-1 mRNA, mapping across the FH mRNA transcript was undertaken by PCR. Results demonstrated approximate equivalence for the levels of transcripts from three sites across the full-length FH mRNA and did not detect FHL-1 or FHR-1 in uninfected or DENV-infected HeLa.
Quantitation of FH protein associated with cell lysates by ELISA, demonstrated an increase in DENV-infected cells. The FH protein, however, was not released from the DENV-infected cell. Immunofluorescent staining demonstrated higher fluorescence intensity for FH on the cell membrane and within DENV-infected compared to uninfected cells, similar to indications of FH on the cell surface of DENV-infected monocyte-derived macrophages, analysed by flow cytometry [10]. The subcellular distribution of FH protein was in-part localised to the plasma membrane, as expected, but was increased in both uninfected bystander and DENV-infected cells in the DENV-challenged population. Additionally, FH showed increased localisation along networks, consistent with the cytoskeleton, and around the nucleus. This perinuclear localisation is reminiscent of the extensive perinuclear hyperproliferation of the endoplasmic reticulum (ER) in DENV-infected cells and may relate to ER-stress [29] and difficulty for the infected cell in producing a large protein such as FH in this context. Intracellular functions of FH have been described in apoptotic cells, where intracellular FH directs cleavage of nascent C3 into C3b, which then generates the opsonin iC3b [30, 31]. Potentially intracellular FH may serve a similar function in virally infected cells. For both membrane, cytoskeletal, and perinuclear associated FH, the increase in immunostaining and altered localisation were observed in cells of the DENV-challenged culture that were undergoing active replication (dsRNA positive) and non-productively infected or uninfected bystander cells (dsRNA negative). This observation is consistent with previous data demonstrating induction of FH in both DENV-infected and uninfected bystander macrophages by flow cytometry, although a bystander effect was not observed in DENV-infected EC [10]. Induction of FH mRNA in trans with > 50 kDa fractionated supernatant from DENV-infected cells was observed, suggesting direct stimulation by a secreted factor. Mass spectrometry analysis of supernatants identified that the > 50 kDa fraction contained DENV structural proteins (envelope and membrane), most likely present as assembled virions and hence retained in the > 50 kDa fraction. The > 50 kDa fraction also contained DENV-NS1, probably as the 310 kDa secreted hexamer [32]. Secreted DENV-NS1 is a potential candidate for FH mRNA induction due to reports that NS1 can activate TLR4 [6, 7]. TLR4 activation in response to lipopolysaccharide (LPS) stimulation is also known to induce FH mRNA in monocytes and macrophages [33]. Blockade of TLR4 by the antagonist TAK-242 during DENV infection of HeLa inhibited DENV-induction of FH mRNA, suggesting that this effect is mediated via TLR4. Stimulation of cells however with supernatant containing NS1 did not induce FH mRNA indicating that NS1 alone cannot mediate this affect. Recent reports have indicated that NS1 functions in binding TLR4 as a multimeric lipoprotein [34]. Thus, to conclusively exclude NS1 as the mediator of DENV-induction of FH, the multimeric forms of NS1 released from cells including as complexes with human lipids, should be investigated.
Although our mass spectrometry analysis was not undertaken in a quantitative manner, the confidence for mass spectrometry identification of some proteins shifted from low-confidence, low-abundance in conditioned media from uninfected cells to high confidence and abundance in the > 50 kDa fraction from DENV-infected cells. These proteins would be interesting targets for future analyses and included C5, FB, C4, alpha2M, and ITIH2. Changes in ITIH2 following DENV infection have not been reported, although there is evidence to suggest in vitro that ITIH2 is necessary for DENV to enter and infect dendritic cells [35]. Interestingly, ITIH2 is considered an acute phase reactant that is induced during inflammation, a serine protease and can bind hyaluronase—a form of GAG, and influence complement, as reviewed [36]. The role of ITIH2 during DENV infection, including the association of ITIH2 with inflammation and ability of ITIH2 to influence GAG levels in and released from DENV-infected cells is of interest. Additionally, MMP2 was uniquely detected in fractionated supernatant from DENV-infected cells. MMP2 is a metalloproteinase that can have a role in extracellular matrix remodelling and may be involved in DENV effects on GAGs or inflammation [37]. The protease inhibitor and chaperonin alpha2M is a member of the complement-coagulation family but also improves DENV virion stability and worsens West Nile virus (WNV) and Japanese encephalitis virus (JEV) neuropathology in mice [38, 39]. Additionally, acutely raised alpha2M is observed in patients who developed dengue with complications compared to dengue [40]. Mass spectrometry data also supported the induction of other complement proteins such as C4, FB, and C5 following DENV infection, as is also documented in dengue patients (C5, FB) and for FB in DENV-infected macrophages and EC [10, 41]. Mass spectrometry data did not show an increase in FH protein in the supernatant, consistent with published data [10] and Figure 1 herein. Thus, the mass spectrometry data here supports investigating further relationships between DENV and these host proteins.
In addition to changes in proteins, DENV infection decreased total cellular and supernatant levels of sulphated GAGs but did not alter levels or binding of FH to cell surface sialic acid. This latter finding is consistent with previous studies in EC [42], although prior reports indicate shedding of sialic acid and GAGs from the glycocalyx of ECs in response to DENV NS1 [42–44]. Although total levels of cell-associated GAGs were reduced by DENV infection, our study demonstrated an increase in the amount of FH that can be released from DENV-infected cells by cleavage with heparinases. Heparan sulphate are important ligands for cell-associated FH, where FH bound extracellularly can regulate factor I-mediated C3 cleavage and complement activity, whilst heparin binding can also facilitate DENV infection of cells [43, 45, 46]. Increased heparan sulphate-bound FH on DENV-infected cells, therefore may be of benefit to the host, where increased heparan sulphate binding by FH may competitively block DENV entry and restrict viral spread [47]. Alternatively, increased cell surface heparan sulphate bound-FH in DENV-infected cells may benefit the virus and protect the infected cells from complement-mediated destruction. The increase in heparan sulphate-FH binding may also account for the discord between increased FH mRNA and unchanged FH secretion from DENV-infected cells, with FH retained at the surface of DENV-infected cells via this ligand. The structural variability of heparan sulphates can impact FH and other FHR-proteins binding to cell surfaces and thus further detail of the heparan sulphate-FH forms bound to DENV-infected cells is warranted to properly understand the implications of this interaction for complement regulation. Additionally, studies such as FH staining of postmortem tissues from fatal dengue cases or mouse models of severe disease would be of interest to assess FH and GAGs in vivo.
In conclusion, DENV-infection influences FH production and biology, an important regulator of the complement AP, in a manner that utilises TLR4 and is secondary to the production of a > 50 kDa soluble factor from infected cells. The DENV-induced FH protein is retained within cells and at the cell surface via binding to heparan sulphate, despite decreased levels of total cell surface GAGs. This occurs in both DENV-infected and uninfected bystander cells, which may have important consequences for blocking DENV entry and restricting spread of infection or protecting infected and uninfected bystander cells from C3b deposition and complement-mediated lysis. This phenomenon appears to be cell-type specific and could be exploited to develop therapeutic strategies for targeting the complement system at the point of FH regulation to decrease C3b and C3a levels and reduce complement-mediated immunopathology.
alpha2M: alpha2-Macroglobulin
AP: alternative pathway
C5: complement component 5
DENV: dengue virus
DMMB: dimethyl methylene blue
EC: endothelial cells
ELISA: enzyme-linked immunosorbent assay
FH: factor H
FHL-1: factor H-like-1
FHR-1: factor H-related protein-1
GAGs: glycosaminoglycans
h: hours
hpi: hours post infection
IFN: type I interferon
ITIH2: inter-alpha-trypsin inhibitor heavy chain H2
kDa: kilodaltons
min: minutes
MOI: multiplicity of infection
NS1: non-structural protein-1
PBS: phosphate buffered saline
SCR1: short consensus repeat 1
TLR4: toll-like receptor-4
Thank you to Dr Sheila Cabezas-Falcon for advice. Thank you to Mr Luke Kris for laboratory assistance.
JGD: Conceptualization, Formal analysis, Investigation, Writing—original draft, Writing—review & editing. BA: Conceptualization, Supervision, Writing—review & editing. EC and VS: Formal analysis, Investigation, Writing—review & editing. DLG: Conceptualization, Funding acquisition, Writing—review & editing. JMC: Conceptualization, Funding acquisition, Supervision, Writing—review & editing. All authors read and approved the submitted version.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
The datasets generated for the mass spectrometry for this study can be found at https://zenodo.org/record/6911339#.YuDXjHZByUl. MP4 digital multimedia images from confocal microscopy are available at https://youtu.be/C7wO1RE_wdw and https://youtu.be/mjwsZh8lshU.
This study was supported by funding from the Flinders Foundation. JGD is supported by a scholarship from the Australian Government Research Training Program. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
© The Author(s) 2024.
Copyright: © The Author(s) 2024. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
