Affiliation:
1College of Biology, Hunan University, Changsha 410082, Hunan, China
ORCID: https://orcid.org/0009-0001-4526-884X
Affiliation:
1College of Biology, Hunan University, Changsha 410082, Hunan, China
ORCID: https://orcid.org/0009-0001-4065-0695
Affiliation:
1College of Biology, Hunan University, Changsha 410082, Hunan, China
ORCID: https://orcid.org/0009-0006-8711-9830
Affiliation:
1College of Biology, Hunan University, Changsha 410082, Hunan, China
ORCID: https://orcid.org/0009-0009-7672-8093
Affiliation:
1College of Biology, Hunan University, Changsha 410082, Hunan, China
ORCID: https://orcid.org/0009-0002-9311-5669
Affiliation:
1College of Biology, Hunan University, Changsha 410082, Hunan, China
ORCID: https://orcid.org/0009-0003-8244-6208
Affiliation:
1College of Biology, Hunan University, Changsha 410082, Hunan, China
ORCID: https://orcid.org/0000-0002-9464-5293
Affiliation:
3Joint Institute of Tobacco and Health, Kunming 650202, Yunnan, China
Email: 125176198@qq.com
Affiliation:
1College of Biology, Hunan University, Changsha 410082, Hunan, China
Email: wanghonghui@hnu.edu.cn
Explor Immunol. 2024;4:533–553 DOI: https://doi.org/10.37349/ei.2024.00157
Received: December 23, 2023 Accepted: July 09, 2024 Published: August 28, 2024
Academic Editor: Dominique J. Charron, Hospital Saint Louis, France
Diabetic wound healing presents a unique and complex challenge due to the impaired cellular and molecular functions associated with diabetes. Chronic wounds in diabetic patients are characterized by prolonged inflammation, reduced angiogenesis, and impaired collagen deposition, which significantly hinder the healing process. This comprehensive review focuses on the innovative applications of designer cytokines in precision therapy for diabetic wound healing, emphasizing the remarkable advancements made in overcoming the limitations of natural cytokines, such as their short half-life, potential cytotoxicity, and lack of specificity. We begin by detailing the intricate biological characteristics of diabetic wounds and the essential role that cytokines play in orchestrating the healing process. The review critically examines the constraints of natural cytokines and traces the evolution of synthetic alternatives, with a particular emphasis on peptide-based and nucleic acid-based artificial cytokines. Advanced strategies for designing these artificial cytokines are discussed, including molecular modifications, functional enhancements, and specificity improvements to better target pathological conditions in diabetic wounds. Furthermore, we explore the utilization of synthetic biology techniques to engineer effective cytokine-based therapies. The promising therapeutic potential of rationally designed cytokines is highlighted, showcasing their ability to modulate the wound microenvironment, enhance tissue regeneration, and reduce chronic inflammation. This review not only provides valuable perspectives on the future research directions but also offers insights into the potential clinical applications of these innovative therapies, aiming to significantly improve the outcomes for patients suffering from diabetic wounds.
Wound healing is a complex and dynamic process that involves various cell types and cytokines, unfolding in a detailed temporal sequence. This process encompasses four integrated and overlapping stages—hemostasis, inflammation, proliferation, and remodeling—which are essential for restoring skin integrity and function [1, 2]. For desirable healing outcomes, these stages must occur in a specific order, at precise times, and with appropriate local concentrations over a fixed duration [3]. Disruption at any stage can lead to delayed healing in acute wounds or chronic non-healing wounds. Particularly, diabetic wounds, characterized by prolonged low-grade inflammation, are an example of such disruption [4]. Diabetic foot ulcers (DFUs), severe complications of diabetes, usually result from neuropathy, peripheral vascular disease, and reduced infection resistance [5]. These chronic wounds frequently lead to amputation and significantly impact the quality of life of patients, also being major causes of hospitalization and mortality among diabetic patients [6, 7].
During hemostasis, platelets release transforming growth factor-β (TGF-β) and other growth factors when they come into contact with injured tissue collagen, initiating clot formation and the creation of a temporary extracellular matrix (ECM) [1]. TGF-β also plays a role in recruiting neutrophils and monocytes, contributing to monocyte-to-neutrophil differentiation [8, 9]. Concurrently, platelets aggregate and adhere to damaged endothelial cells, triggering clotting and converting fibrinogen into fibrin, leading to the formation of a clot and a temporary ECM [10]. In the inflammatory phase, pro-inflammatory or activated macrophages play a crucial role in producing and releasing cytokines, including interleukin-1β (IL-1β), tumor necrosis factor-α (TNF-α), IL-6, and IL-8 [11]. This phase is characterized by increased inflammatory cell infiltration in the dermis and blood vessel walls, resulting in elevated levels of cytokines such as IL-6, IL-1β, and TNF-α, ultimately leading to a reduction in the expression of growth factors like TGF-β [12]. Furthermore, immune cells can trigger the release of cytokines like IL-1β, IL-6, and IL-23 [13]. IL-1β activation leads to lysosomal secretion, caspase-1 cleavage, increased insulin resistance, and delayed wound healing [14]. TNF-α has a dual role, initiating inflammation at low concentrations and activating MAPK and NF-κB signaling pathways through binding with TNF receptor-2 (TNFR-2) at high concentrations, thereby promoting wound healing [15]. IL-6 stimulates B lymphocyte growth, neutrophil production in the bone marrow, and the release of acute phase proteins in the liver [16]. It may also influence leukocyte recruitment by inducing endothelial cells to secrete IL-8 and monocyte chemoattractant protein-1 (MCP-1) [17]. Additionally, IL-1β and IL-23 promote the release of IL-17 [18], which facilitates blood cell infiltration and creates an inflammatory environment conducive to wound healing [19]. During the proliferation phase of diabetic wound healing, several key cytokines play critical roles in the transition from inflammation to tissue repair. TGF-β, IL-10, hypoxia-inducible factor-1α (HIF-1α), and peroxisome proliferator-activated receptor-γ (PPAR-γ) are involved in facilitating the shift from pro-inflammatory to anti-inflammatory macrophages [20]. In hypoxic conditions, HIF-1α stimulates the formation of new blood vessels and upregulates genes associated with wound repair, including vascular endothelial growth factor (VEGF) and adhesive integrins [21]. Anti-inflammatory macrophages release various factors, including IL-1β, matrix metalloproteinase-9 (MMP-9), IL-10, IL-6, and TGF-β, and they express high levels of inducible nitric oxide synthase (iNOS). These factors contribute to the remodeling of blood vessels and the ECM while helping to reduce inflammation [22]. In the final remodeling phase of the healing process, granulation tissue is gradually replaced by scar tissue, contributing to the restoration of tissue integrity and function.
In the treatment of diabetic wounds, cytokines are considered a promising therapeutic agent. However, in diabetic wounds, immune regulation is impaired, and inflammation persists, limiting the therapeutic efficacy of cytokines. The majority of cytokines have a short half-life and low stability, making it challenging for them to sustain their effects in wounds. Therefore, to maintain the bioactivity of cytokines and achieve sustained release, appropriate wound dressings are needed to more effectively utilize cytokines in promoting the healing of diabetic wounds. For instance, Wang et al. [21] developed a novel wound dressing embedding IL-33 in a DNA hydrogel, and the results showed that local inflammation in diabetic wounds was addressed and granulation tissue regeneration improved, leading to accelerated wound healing through IL-33-cycle treatment. Wound dressings based on skin substitute therapy are one of the most promising methods for the treatment of diabetic wounds. For example, Liu et al. [22] have developed various functionalized hydrogels for diabetic wound treatment. The research group reported a new sodium alginate/chitosan (SA/CS) Janus hydrogel dressing in 23 years, which can prevent infections and achieve sustained release of l-arginine to promote blood vessel regeneration. In addition, a water-based gel based on okra was studied, which can promote cell migration, blood vessel formation, and re-epithelialization of chronic wounds in diabetic rats, significantly promoting wound remodeling [23]. This year, a new bioactive hydrogel containing MAP (magnesium ascorbyl phosphate) encapsulated in microspheres has been reported. The experimental results show that these wound dressings significantly promote the healing of diabetic wounds through multiple biological effects [24].
Cells play a vital role in the healing of wounds, with chronic wounds like DFUs compromised in healing due to abnormal behavior of various cells. Cell therapy can correct factors leading to prolonged wound healing through various mechanisms. For instance, mesenchymal stem cells (MSCs) promote the healing process by shortening the inflammatory phase, regulating cytokine levels, and enhancing angiogenesis. Studies in different animal models indicate that MSCs can decrease the number of inflammatory cells, reduce levels of pro-inflammatory cytokines (IL-1, IL-6, TNF-α), increase levels of anti-inflammatory factors (IL-10 and VEGF), thus aiding in reducing scar formation and promoting wound healing. Additionally, MSCs influence gene expression, enhance collagen content, and reduce MMP-9 levels. MSCs also can secrete VEGF to promote angiogenesis and differentiation of endothelial progenitor cells into endothelial cells. Bone marrow mononuclear cells (BM-MNCs) and peripheral blood mononuclear cells (PB-MNCs) have functions such as promoting angiogenesis, reducing inflammation, enhancing re-epithelialization, and increasing collagen deposition. They secrete angiogenic factors [VEGF, basic fibroblast growth factor (bFGF), Ang1] and anti-inflammatory factors, aiding in wound healing.
Moreover, fibroblasts can secrete growth factors and components of the ECM to promote wound healing. Keratinocytes promote wound healing by expressing growth factors [such as VEGF, bFGF, platelet-derived growth factor (PDGF)], and ECM proteins (fibronectin, collagen, etc.). Currently, cell therapy as a novel treatment for diabetic ulcers is gaining increasing attention, with its effectiveness being confirmed in clinical trials. The steady development in synthetic biology enables scientists to use genetically engineered cells as the basis for developing new therapies instead of small molecules or biological agents. Cells equipped with synthetic gene circuits can control the targeting, timing, and dosage of therapeutic activities based on specific disease biomarkers, becoming a powerful new weapon against diseases.
Furthermore, the use of synthetic gene circuits may be a key feature of advanced cell therapy, addressing some significant limitations of traditional therapies, namely lack of flexibility, specificity, and predictability [25]. The current application of some cell therapies in diabetic wound healing has entered clinical trials. However, cell therapy has some inevitable defects, such as high cost, safety risks, low survival rate, and difficult-to-control differentiation problems, significantly limiting the role of cells in diabetic wound healing. In contrast, cytokine-enabled therapy has high efficiency and wide safety advantages, so we are more focused on the design of the engineered cytokines. In the context of diabetic wound healing, cytokines play a pivotal role in regulating various aspects of the healing process [2, 16, 26–35] (Table 1). However, natural cytokines have their limitations, including short half-lives, unintended side effects, and pleiotropic actions, which can hinder their effectiveness [36]. Consequently, relying solely on natural cytokines may not provide optimal outcomes for diabetic wound healing. In recent years, researchers have begun to explore the use of engineered cells and cytokines to promote diabetic wound healing. The importance of engineered cell factors in diabetic wound healing cannot be ignored. They can help improve the speed and quality of wound healing in diabetic patients, reducing the risk of infection and complications. Recent advancements in research have illuminated the potential of synthetic cytokines as a solution to these limitations. Synthetic cytokines can be engineered to address specific therapeutic needs, offering greater control and precision in modulating the healing response. This review explores the development strategies behind synthetic cytokines, delving into the principles of their design and engineering (Figure 1). Additionally, it examines their diverse applications in the treatment of various diseases, highlighting their potential to revolutionize diabetic wound healing by enhancing the body’s natural regenerative processes.
Different cell types secrete different cytokines at different stages of wound healing
| Phase in wound healing | Cytokine | Cell type | Reference(s) |
|---|---|---|---|
| Hemostasis | PDGF, TGF-β, IGF | Platelet | [2, 26, 27] |
| Inflammation | IL-1β, IL-6, IL-8, IL-23, TNF-α, IFN-γ | M1 macrophages | [28, 29] |
| Inflammation | TGF-β, IL-4, IL-5, IL-6, IL-10, IL-13 | T lymphocytes | [16, 26, 30, 31] |
| Proliferation | IL-10, VEGF, TGF-β, FGF, EGF, MMP-9 | M2 macrophages | [28, 32, 33] |
| Proliferation remodeling | MMPs, KGF, IL-6, IGF | Fibroblast | [27, 34] |
| Proliferation remodeling | VEGF, TGF-α | Malpighian cell | [26, 35] |
PDGF: platelet-derived growth factor; TGF: transforming growth factor; IFN-γ: interferon-γ; FGF: fibroblast growth factor; KGF: keratinocyte growth factor
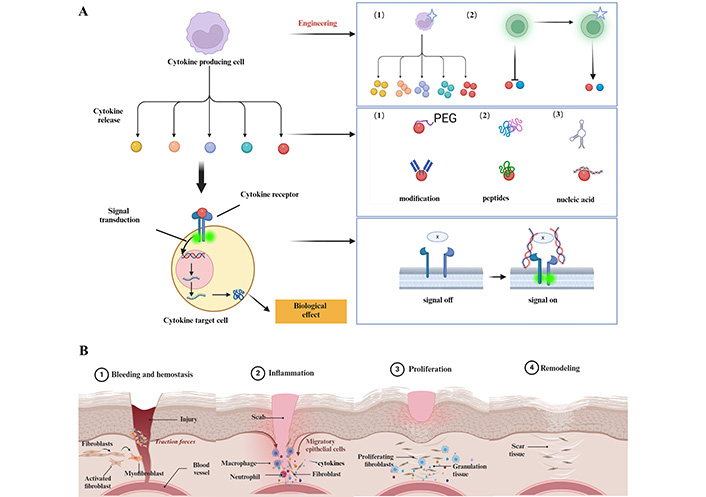
Design strategy for synthetic cytokines and schematic representation of diabetic wound healing. (A) It is possible to design and synthesize artificial cell factors from different perspectives such as cells, cell factors, and receptors. This includes generating more cell factors through cell modification or modifying cells to produce cell factors that were not produced before. It also involves modifying natural cell factors and synthesizing cell factors based on peptides and nucleic acids, as well as modifying receptors to change their affinity for binding cell factors; (B) Diabetes wound healing can be divided into four stages: hemostasis, inflammation, proliferation, and remodeling. Each stage involves complex regulation with the participation of various cells and cytokines. Created with BioRender.com
Cytokines, which play a pivotal role in human physiology and pathology, coordinate a complex signaling network with diverse activities and pluripotent functions. Despite their numerous benefits, cytokine therapy has faced challenges characterized by side effects and a limited therapeutic range, historically limiting the application of natural cytokines as pharmaceutical agents. However, as our understanding of the structural principles governing cytokine-receptor interactions and functional signaling deepens, novel approaches in protein engineering and synthetic design have emerged. These innovative methods provide practical and potent means for artificially modifying natural cytokines [37]. Recent advancements in this field have a specific focus on enhancing the therapeutic potential of cytokines through various strategies, including domain modification, extending their half-life, creating fusion proteins, and developing dual-function fusion cytokines. These cutting-edge approaches hold promise for overcoming the historical constraints of cytokine therapy and expanding their utility as therapeutic agents in medicine.
Generally, natural cytokines have encountered limitations in disease treatment, primarily due to their varying affinities for receptors and other cytokines, which can diminish their therapeutic efficacy. In response, recent research efforts have focused on the modification and optimization of natural cytokines to enhance their effectiveness. Activation of receptors often triggers a series of conformational changes and autophosphorylation events. The conformational changes of the receptor can influence signal transduction. Therefore, through studying the structure of cytokine-receptor complexes, we can guide the rational design of cytokines. Kim et al. [38] utilized techniques such as cryo-electron microscopy to investigate the structures of the insulin receptor (IR) and various nucleic acid insulin analogs. Analysis of these structures revealed their roles in IR activation, phosphorylation processes, and the mechanism of selective activation, providing a structural basis for designing selective agonists of IR. Thus, we can determine the structure of ligands, receptors, or ligand-receptor complexes through methods like cryo-electron microscopy and X-ray crystallography and then carry out engineering modifications guided by these structures. Understanding how ligands and receptors bind and the subsequent signal transduction process is crucial. By selecting the structural interface based on different needs, we can enhance or weaken the affinity between a specific ligand and receptor through structural domain modifications. Furthermore, the structural design of ligand mimetics can be developed to substitute ligands and exert their functions.
For instance, consider the case of IL-2, a cytokine responsible for stimulating the proliferation and effector function of NK cells and T cells. Although recombinant IL-2 has shown promise in treating diseases like metastatic liver cancer, its clinical application is hindered by potential severe side effects, including pulmonary edema, effector cell-induced cell death, and the expansion of regulatory T cells (Tregs) [39, 40]. The IL-2 receptor comprises three subunits: α (CD25), β (CD122), and γ (CD132). The α subunit exhibits low affinity and lacks downstream signal transduction, while the β and γ subunits, when combined, form a receptor with moderate affinity that triggers downstream signaling via Janus kinases (JAK). When the α subunit is present, the heterotrimer forms a high-affinity receptor [41, 42]. Researchers have harnessed mutations and molecular modifications to alter IL-2’s affinity for different receptor subunits, thereby enhancing its efficacy and reducing toxicity. Levin et al. [43] have developed an IL-2 super-agonist that promotes NK cell and CD8+ T cell proliferation while minimizing Treg expansion and the occurrence of pulmonary edema. Other companies are also designing IL-2 super agonists for various indications, such as MDNA209, which is utilized to inhibit abnormal T cell function [44].
IL-4, another significant cytokine produced by helper T cells, plays a role in guiding CD4+ T cell differentiation and antibody production. However, its therapeutic use has been limited due to associated toxicity. Researchers, including Junttila et al. [45], have developed two types of IL-4 super-agonists that selectively activate different cell receptors, aiming to reduce toxic side effects. Furthermore, modifications to the binding domains of other cytokines have resulted in the creation of super-agonists or antagonists capable of regulating downstream signaling pathways. For instance, Glassman et al. [46] designed a selective partial agonist of IL-12, Medicenna developed two IL-13 super-agonists, MDNA132 and MDNA413, and in recent years, Yin et al. [47] and Junttila et al. [48] have designed different IL-10 mutants. Together, the natural cytokine can be modified and optimized to improve their therapeutic efficacy while reducing toxic side effects. These engineered cytokines, including super-agonists and receptor-selective variants, offer enhanced treatment options for a range of diseases. The structural modifications of natural cytokines have paved the way for more effective and targeted therapies, marking a significant advancement in the field of cytokine-based treatments (Figure 2A). Furthermore, de novo protein-based factors can be rationally designed through protein structure prediction. Zhu and colleagues [49] have developed a novel machine learning-based virtual screening approach for various PI3Kc protein structures, leading to the discovery of new PI3Kc inhibitors. This approach also offers new insights for enhancing cytokine stability and receptor interactions.
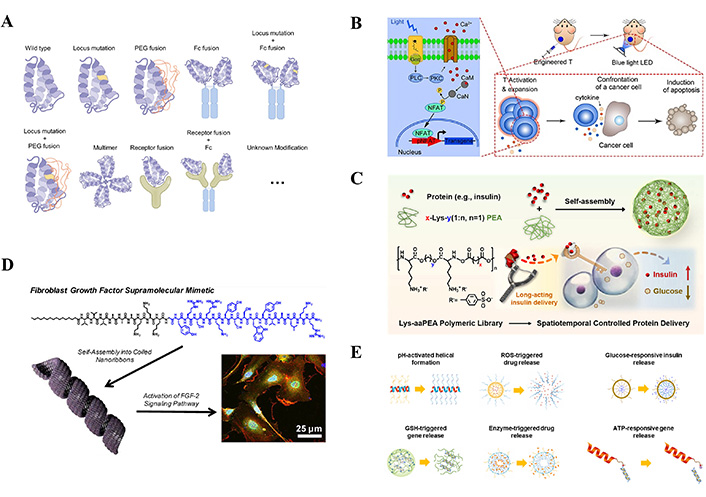
Engineering approaches to modify the therapeutic potency of cytokines and construction strategies based on polypeptide synthetic cytokines. (A) Natural cytokine modification method includes site mutation, PEG modification, fusion protein, etc. (B) The engineered T cells carrying the transgenic “payload” remodel the tumor microenvironment by remotely controlling the concentration of transgenic cytokines such as IL-2, IL-15, and TNF-α. (C) Lys-aaPEAs can interact with and encapsulate proteins into nanocomplexes via electrostatic interactions, which can be used to spatiotemporally control protein delivery. (D) Cytokine analog peptide. (E) The responsiveness of the polypeptide is customized by introducing ideal functional fragments responding to specific stimuli, with pH response, ROS response, glucose response, GSH response, enzyme response, and ATP response, etc.
Note. Figure A reprinted with permission from “The use of supercytokines, immunocytokines, engager cytokines, and other synthetic cytokines in immunotherapy” by Zheng X, Wu Y, Bi J, Huang Y, Cheng Y, Li Y, et al. Cell Mol Immunol. 2022;19:192–209 (https://www.nature.com/articles/s41423-021-00786-6). © 2022 Springer Nature. Figure B reprinted from “An Optogenetic Controllable T Cell System for Hepatocellular Carcinoma Immunotherapy” by Zhao B, Wang Y, Tan X, Zheng X, Wang F, Ke K, et al. Theranostics. 2019;9:1837–50 (https://www.thno.org/v09p1837.htm). CC BY-NC. Figure C reprinted with permission from “Development of a Lysine-Based Poly(ester amide) Library with High Biosafety and a Finely Tunable Structure for Spatiotemporal-Controlled Protein Delivery” by Han S, Wu J. ACS Appl Mater Interfaces. 2022;14:55944–56 (https://pubs.acs.org/doi/10.1021/acsami.2c16492). © 2022 American Chemical Society. Figure D reprinted with permission from “Mimicking the Bioactivity of Fibroblast Growth Factor-2 Using Supramolecular Nanoribbons” by Rubert Pérez CM, Álvarez Z, Chen F, Aytun T, Stupp SI. ACS Biomater Sci Eng. 2017;3:2166–75 (https://pubs.acs.org/doi/10.1021/acsbiomaterials.7b00347). © 2017 American Chemical Society. Figure E reprinted from “Stimuli-Responsive Polypeptides for Biomedical Applications” by Lee D, Rejinold NS, Jeong SD, Kim YC. Polymers (Basel). 2018;10:830 (https://www.mdpi.com/2073-4360/10/8/830). CC BY.
The therapeutic potential of recombinant cytokines is often limited by their short serum half-life. To address this challenge, extensive effort has been made to explore various methods for extending cytokines’ half-life and enhancing their biological activity. These strategies encompass chemical modifications such as PEGylation, lipidation, genetic engineering, and disulfide bond formation (Figure 2A). Notably, innovative approaches have yielded promising results. Recombinant PEG-IL-2 and pegylated Fc-IL-2, for instance, have demonstrated prolonged plasma clearance and notable anti-tumor activity [50]. Furthermore, advanced IL-2 variants, including NKTR-214 and AMG592, have been engineered to possess extended half-lives and selective activity [44, 51]. Emerging candidates like NKTR-358 and MDNA11 exhibit enduring high-affinity characteristics, holding significant potential in anti-tumor therapy and clinical trials, particularly for chronic graft-versus-host disease and cancer treatment [52]. Genetic modifications have also been explored to enhance cytokine thermal stability. For example, the introduction of triple mutations into FGF-1, augmenting van der Waals forces and steric hindrance resulted in a remarkable 21.5°C increase in denaturation temperature compared to wild-type FGF-1 [53, 54]. Moreover, addressing protein proteolytic resistance, the introduction of two mutations at a known protease cleavage site in FGF-1 led to a substantial up to 100-fold increase [55]. The incorporation of disulfide bonds, pivotal components of protein structure, has shown great potential for enhancing stability and thereby promoting biological activity [56]. A notable illustration involves site-specific mutagenesis, where replacing Ala66 in FGF-1 with cysteine introduced a disulfide bond, resulting in a remarkable 14-fold increase in the variants’ half-life [57]. Therefore, various strategies have been developed to enhance the stability and activity of natural cytokines, demonstrating promise in treating a wide range of diseases. With ongoing research and clinical trials, the development of novel cytokine-based therapies holds the potential to transform the landscape of medical treatments, offering improved outcomes for patients.
Due to the severe toxic side effects and poor pharmacokinetic characteristics exhibited by the direct use of cytokines for treatment, a series of methods have been designed to reduce these side effects, including the construction of cytokine fusion proteins (Figure 2A). Fusion proteins are proteins composed of at least two structural domains, which are encoded by independent genes that are connected. Therefore, they are transcribed and translated into a single unit, resulting in a single polypeptide. Moreover, cytokine fusion proteins have one of their structural domains derived from cytokines. Depending on the source of the other structural domain of the fusion protein, it can be classified as immune cytokines, bispecific cytokine fusion proteins, or chimeric cytokines. Immunocytokines are produced by fusing antigen-specific antibodies with immune-enhancing cytokine genes [58]. Immunocytokines can be further classified based on their structure, including fusion proteins of cytokines fused with immunoglobulins, fusion proteins of cytokines fused with antibody Fc fragments, and fusion proteins of cytokines fused with antigen-binding fragments of antibodies such as Fab fragments, single-chain variable fragments (scFv), or bivalent derivatives. Furthermore, bispecific cytokine fusion proteins are formed by fusing two cytokines with antibodies, such as the fusion protein targeting IL-2-F8-TNFmut constructed by De Luca and colleagues [59]. This fusion protein of dual cytokines exhibits in vivo anti-tumor activity in immunocompetent mouse tumor models WEHI-164, CT26, LLC, and F9 teratoma, whether used alone or in combination with mouse PD-L1 specific monoclonal antibodies.
Chimeric cytokines refer to fusion proteins composed of functional factors and cytokines. Penafuerte et al. [60] designed a fusion protein of granulocyte-colony stimulating factor and IL-2 called “hGIFT2”. hGIFT2 possesses unique biochemical characteristics distinct from IL-2 and granulocyte-colony stimulating factors and is an effective tool for activating and maturing NK cells, potentially applicable for tumor immunotherapy. Additionally, chimeric cytokines can also be fused with antibodies, the LIF receptor (LIFR) domain binding ciliary neurotrophic factor (CNTF) was replaced with IL-6 to form IC7, and then the Fc domain of IgG was fused with IC7 to obtain IC7Fc. Intraperitoneal injection of IC7Fc significantly reduced overall body weight and fat weight in mouse models. Furthermore, IC7Fc can lower fasting blood glucose levels and enhance glucose tolerance. These results suggest that IC7Fc can improve physiological indicators. Findeisen et al. [61] are actively promoting phase I clinical trials of IC7Fc, believing it to be a promising drug for the future treatment of type 2 diabetes. Hsu et al. [62] engineered IL-12 as a cytokine by attaching it to a cleavable tumor protease binding site, allowing for selective cleavage and activation of the immune response. This approach presents an effective tumor-targeted therapeutic strategy with reduced toxicity. On the other hand, Mansurov et al. [63] proposed a protein engineering solution to IL-12 toxicity by fusing a domain of the IL-12 receptor to IL-12 using a tumor-associated protease cleavable linker, significantly restraining the pro-inflammatory effects of IL-12 at tumor sites.
In the realm of modern medicine, the role of synthetic biology in engineering cells capable of secreting cytokines has become increasingly significant. Traditional recombinant cytokines, which simply replace the endogenous cytokines of cells, often lead to reduced efficacy and specificity. However, recent technological advancements have spotlighted engineered cells as a groundbreaking approach. These cells can be genetically altered to control the synthesis and release of cytokines, allowing for targeted intervention in specific disease environments (Figure 2B). Interestingly, Zhao et al. [64] designed a rationally engineered T cell carrying a transgenic “payload”. This innovation enables the reshaping of the tumor microenvironment through remote control of the concentration of transgenic cytokines such as IL-2, IL-15, and TNF-α. The precise regulation of these cytokines’ release bolsters the proliferation and persistence of T cells within the tumor, thereby augmenting the cytolytic effectiveness of the engineered cells.
The functional efficacy of cytokines is intrinsically linked to the molecular recognition and signal transduction between the cytokines and surface receptors on effector cells. Consequently, the reconstruction or reprogramming of this molecular interaction presents a promising avenue for novel disease treatment methodologies. Inspired by chimeric antigen receptor (CAR) technology, the field of receptor reprogramming has garnered extensive interest. Modifying the molecular recognition units of receptors could yield cell membrane receptors with enhanced specificity and affinity. This approach leverages genetic engineering techniques to redesign the cytokine-receptor interactions, activating downstream signaling pathways crucial in numerous biological functions, including development, wound healing, and immune responses. In 2017, the Schwarz group [65] introduced a modular extracellular sensor architecture (MESA) modification for immune T cells. This modification endowed the T cells with a VEGF-specific response, enhancing IL-2 secretion and bolstering the immune response. Following this, in 2018, the Chang research group [66] reported the development of a TGF-β responsive CRA immune T cell (TGF-β-CAR-T). This cell type engages in signaling via the specific interaction with TGF-β, thereby promoting T cell proliferation and cytotoxicity. This process effectively converts the immunosuppressive attributes of TGF-β into a mechanism for immune activation. The approval of CAR-T cells has opened the door to synthetic biology genetic cell therapies. However, the production of patient-specific transgenic T cells still poses challenges that need to be addressed due to time-consuming and expensive issues. The field of engineered cell therapy is gradually maturing but faces unprecedented challenges in manufacturing and clinical development as cells have not yet been widely used as the basic unit for in vivo production of therapeutic drugs. Some of the technical challenges in manufacturing include the accumulation of mutations during the production process, as well as the emergence of variant strains due to production load and metabolic burden, which may make consistently producing approved batches challenging. In addition, clinical development faces challenges such as patient skepticism [67].
Together, synthetic biology has become crucial in modern medicine for its role in engineering cells to secrete specific cytokines, enhancing disease targeting and treatment efficacy. This approach, advancing beyond traditional methods, involves genetically modifying cells to precisely control cytokine release, exemplified by notable achievements like engineered T cells for tumor therapy and receptor reprogramming techniques. These innovations highlight synthetic biology’s significant impact on developing new medical therapies.
Cytokines are bioactive proteins that have important implications in treating various critical human diseases based on protein-based therapy. However, the fragile structure of proteins, susceptibility to enzymatic degradation, poor stability, and propensity to trigger immune responses pose limitations to protein therapy in certain cases. Additionally, in the development of protein therapies targeting intracellular targets, limitations in membrane permeability and endosomal escape could affect protein internalization and biological activity. Therefore, developing effective protein delivery strategies is crucial to enhance therapeutic efficacy and enable widespread medical applications [68]. Traditional protein delivery strategies often face issues such as unpredictability, low bioavailability, and lack of control. Nowadays, nanotechnology is being utilized to address these challenges encountered in traditional protein delivery. For instance, Han and Wu [69] reported a new versatile protein delivery nano-platform based on lysine—Lys-aaPEA protein delivery system (Figure 2C). Insulin was chosen as the model protein to evaluate the protein delivery effectiveness. Experimental results indicate that with this method, insulin can successfully exert its biological effects. Additionally, subcutaneous injection of Lys-aaPEA and insulin nanocomplex (Lys-aaPEA@INS) in chemically induced type I diabetes mouse model demonstrated long-term hypoglycemic activity, further confirming the feasibility of such nanoparticle carriers for spatiotemporal insulin delivery in vivo. Building on the work mentioned above, Han and colleagues [70] have designed a family of poly(ester amide)s based on lysine, known as Lys-aaPEAs, as a versatile oral protein delivery platform for efficient protein loading and degradation prevention. When decorated with hyaluronic acid (HA) to carry insulin, Lys-aaPEAs exhibit good oral hypoglycemic effects in type 1 diabetic mice and reduce complications. Although nanotechnology shows promise in the field of protein therapy, nano-sized protein delivery systems are still in the early stages of clinical development and face multiple challenges. These include the need for precise drug encapsulation, achieving controlled release and intelligent design, simplifying production scale-up, enhancing intracellular delivery efficiency, and enabling targeted therapy and personalized medicine while also requiring a comprehensive assessment of safety and long-term performance [68].
Peptides, as biopolymers, are composed by linking amino acid residues through peptide bonds. They exhibit a range of beneficial features, including high biological activity, biocompatibility, small molecular weight, good permeability, and exceptional targeting abilities [71]. Their adjustable amino acid structures and sequences enable flexible design and customization, facilitating the formation of specific biomimetic, recognition, and targeting properties through molecular self-assembly into supramolecular nanoassemblies. These properties of peptide nanoassemblies are increasingly utilized in biomedical and tissue engineering applications. In comparison to full-length proteins, peptides offer advantages such as reduced degradation, prolonged functionality, higher purity, and lower sensitivity to pH and temperature variations. In some instances, peptides can also achieve more precise cell signal transduction [72].
Recent research has focused on leveraging these peptide advantages for constructing cell factors. Functional peptides have been developed that can either fully substitute for cell factors or regulate their interactions with receptors. Proteins consist of amino acid chains called peptides, containing biologically active sequences known as epitopes. These epitopes, short amino acid sequences, are vital for the “functional coding language” of proteins. They can mimic cytokines and growth factors to specifically bind receptors, initiating downstream cascade reactions, influencing transcription, and playing roles in stem cell phenotype, embryonic development, tissue homeostasis, and repair processes. For example, the mimetic peptide LIANAK of TGF-β1, when coupled with self-assembling RADA16 hydrogels, enhances the expression of cartilage genes and ECM deposition [73]. The peptide KLTWQELYQLKYKGI (KLT), which mimics VEGF, binds to VEGF receptors to activate cell signaling and promote angiogenesis [74]. The mimetic peptide YRSRKYSSWYVALKR of FGF-2 is used in vascularization peptide nanobelts hydrogels, stimulating proliferation and migration of human umbilical vein endothelial cells in vitro [75], and aiding in angiogenesis and wound healing (Figure 2D). Peptides derived from NGF and BDNF, like CTDIKGKCTGACDGKQC and RGIDKRHWNSQ, have been shown to promote neuronal growth and exhibit neuroprotective effects akin to full-length growth factors [76, 77]. Furthermore, growth factor-binding peptides, viewed from the perspective of cell receptors, can regulate the affinity and specificity of growth factor binding to receptors. For instance, heparin peptide nanofibers, designed to mimic heparin’s properties, can bind to various growth factors, inducing angiogenesis both in vitro and in vivo [78]. Together, the exploration of peptide-based approaches opens new avenues for medical research and treatment, highlighting the potential of peptides in replicating and enhancing the functions of traditional growth factors and cytokines.
Peptides can be engineered with selective responsiveness to physical or chemical stimuli and have emerged as promising candidates for “smart” drug delivery systems. Stimuli-responsive polymers are materials that react to specific triggers such as pH, temperature, redox potential, light, and enzymes (Figure 2E). This reaction results in changes in their chemical or physical properties [79]. These polymers are particularly advantageous in biomedical applications because their response is localized, occurring only at the site of the lesion. This localization induces desired therapeutic effects while minimizing side effects [79]. Among stimuli-responsive materials, peptides hold a special place due to their inherent controllability and ease of synthesis. Certain amino acids, like cysteine, glutamic acid, aspartic acid, histidine, and tyrosine, naturally exhibit stimulus-responsive characteristics. This strategy allows for the synthesis of responsive peptides from natural amino acid residues without the need for complex modifications [80, 81]. However, to achieve more complex functionalities, peptides often require coupling methods that attach necessary functional groups or building blocks to their side chains [82].
One area of active research is the development of pH-responsive peptides. These peptides can undergo conformational changes under varying pH conditions, which in turn affects their cell-penetrating abilities. Notable examples include poly(4-(3-acrylic acid)benzoyl-L-lysine) (PABL3) and RP4F (a pH-activated mitochondria-destabilizing helical polypep-tide)[83, 84]. These peptides form helical structures in low pH environments and have shown the ability to selectively penetrate cancer cells. This characteristic suggests their potential use in targeting tumors in vivo and inhibiting tumor growth, offering a promising new direction in cancer therapy. In 2022, the Dong research group [85] reported a significant development in this field. They designed a peptide-rich in thiols that, in an oxidizing environment, folds into an amphiphilic β-hairpin conformation through the formation of two hetero-disulfide bonds. The folding event triggers the self-assembly of the peptide into a mechanically rigid hydrogel. This innovative approach, utilizing multiple disulfide bonds to respond to an oxidizing environment, marked the first instance of controlling conformational changes and self-assembly in this manner.
Moreover, ionic peptides such as poly(lactic-co-glycolic acid) (PLGA) and poly-L-lysine (PLL) are particularly effective in this role. They can physically bind with drugs carrying an opposite charge, forming complexes that are sensitive to pH changes in the environment. When exposed to varying pH levels, these peptide-drug complexes undergo gradual changes. The binding between the peptide and the drug weakens, leading to the controlled release of the drug [86–89]. Moreover, the conformation of the peptide itself alters in response to pH changes, further facilitating drug release. This dual mechanism—the weakening of physical binding and conformational changes in the peptide—allows for precise control over the rate and timing of drug release. This modality represents an intelligent approach to drug delivery, offering significant benefits in terms of treatment efficacy and patient compliance. In addition to physical binding mechanisms, peptides are versatile in their chemical functionality. They contain various functional groups that enable the formation of chemically unstable bonds with drugs. This strategy allows for the loading of drugs within peptide structures through these labile bonds. Upon exposure to specific triggers, such as enzymatic activity or changes in the cellular environment, these unstable bonds break, releasing the drug at the desired site and time [90–92].
Taken together, peptide-based cytokine engineering is a transformative field in biomedical research, harnessing the unique properties of specific amino acids for targeted cancer therapy and responsive drug delivery. Engineered peptides demonstrate the ability to alter their structure in response to environmental stimuli, such as pH changes, facilitating targeted drug release and enhanced cell manipulation. Therefore, the peptide-based artificial cytokine represents a promising direction for developing advanced, efficient therapeutic strategies in medicine.
The unique properties of nucleic acid molecules, including precise complementary pairing ability, sequence programmability, structural diversity, and mechanical properties, make them exceptional nanomaterials for constructing molecular machines [93]. In recent years, the rapid advancement of DNA nanotechnology has led to the widespread use of DNA molecular machines in various fields, including chemistry, materials science, environmental science, and biomedicine [94]. Compared to recombinant proteins, nucleic acid aptamers offer several advantages, including predictable secondary structures and high thermal stability, which reduce the risk of thermal denaturation and variability between ligand batches. These characteristics position DNA aptamers as ideal synthetic alternatives to recombinant growth factors and cytokines. Over the past few decades, researchers have focused on elucidating the principles of biomolecular design and have successfully developed numerous nucleic acid aptamers as modulators of cell signaling induced by mimics of cell factors or as modulators of cell surface receptor binding to cell factors. For instance, in 2019, the Ueki group [95] reported a functional mimic of bFGF composed of only 76 single-stranded DNA chains. This mimic effectively supports the self-renewal and pluripotency induction of induced pluripotent stem cells (iPSCs). Furthermore, they have developed a hepatocyte growth factor mimic capable of inducing growth factor receptor dimerization and mitigating the progression of Fas-induced fulminant hepatitis in a mouse model [96] (Figure 3A).
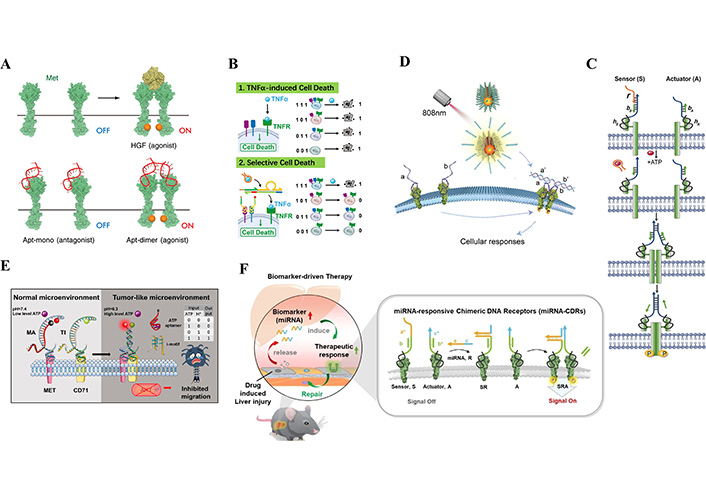
Methods based on the synthesis of cytokines using nucleic acids. (A) Hepatocyte growth factor mimics dimerize growth factor receptors. (B) A DNA automaton that interacts with TNF-α and its receptor TNFR, recognizes and regulates programmed death of target cells in a multicellular environment. (C) DNA agonist construction strategies for growth factor receptors that activate cell signaling by chemical or light control of receptor dimerization. (D) DNA agonist construction strategies for growth factor receptors that activate cell signaling by chemical or light control of receptor dimerization. (E) DNA nanodevices that are responsive to multiple environmental inputs like pH and ATP. (F) A modular and programmable miRNA response to a chimeric DNA receptor (miRNA-CDR) combines miRNA responsiveness with chemical operational surface receptors to initiate receptor dimerization to open downstream signaling pathways
Note. Figure A adapted from “A chemically unmodified agonistic DNA with growth factor functionality for in vivo therapeutic application” by Ueki R, Uchida S, Kanda N, Yamada N, Ueki A, Akiyama M, et al. Sci Adv. 2020;6:eaay2801 (https://www.science.org/doi/10.1126/sciadv.aay2801). CC BY-NC. Figure B adapted with permission from “Scan and Unlock: A Programmable DNA Molecular Automaton for Cell-Selective Activation of Ligand-Based Signaling” by Zhang J, Qiu Z, Fan J, He F, Kang W, Yang S, et al. Angew Chem Int Ed Engl. 2021;60:6733–43 (https://onlinelibrary.wiley.com/doi/10.1002/anie.202015129). © 2020 Wiley-VCH GmbH. Figure C reprinted with permission from “A DNA-Mediated Chemically Induced Dimerization (D-CID) Nanodevice for Nongenetic Receptor Engineering To Control Cell Behavior” by Li H, Wang M, Shi T, Yang S, Zhang J, Wang HH, et al. Angew Chem Int Ed Engl. 2018;57:10226–30 (https://onlinelibrary.wiley.com/doi/10.1002/anie.201806155). © 2018 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. Figure D reprinted with permission from “Near-Infrared Light-Activated DNA-Agonist Nanodevice for Nongenetically and Remotely Controlled Cellular Signaling and Behaviors in Live Animals” by Wang M, He F, Li H, Yang S, Zhang J, Ghosh P, et al. Nano Lett. 2019;19:2603–13 (https://pubs.acs.org/doi/10.1021/acs.nanolett.9b00421). © 2019 American Chemical Society. Figure E reprinted with permission from “Logic Nanodevice-Mediated Receptor Assembly for Nongenetic Regulation of Cell Behavior in Tumor-like Microenvironment” by Wu Y, Huang J, He H, Wang M, Yin G, Qi L, et al. Nano Lett. 2023;23:1801–9 (https://pubs.acs.org/doi/10.1021/acs.nanolett.2c04657). © 2023 American Chemical Society. Figure F reprinted with permission from “An Extracellular miRNA-Responsive Artificial Receptor via Dynamic DNA Nano-assembly for Biomarker-Driven Therapy” by He F, Wang M, Wang J, Wang HH, Nie Z. Angew Chem Int Ed Engl. 2023;62:e202305227 (https://onlinelibrary.wiley.com/doi/10.1002/anie.202305227). © 2023 Wiley-VCH GmbH.
The progress in functional nucleic acid-based molecular recognition, exemplified by nucleic acid aptamers and nucleases, along with advancements in DNA nanostructure construction and dynamic regulation techniques, has laid the groundwork for the development of customizable DNA molecular machines with specific biological functions. These nucleic acid-based molecular tools offer distinct advantages for the construction and regulation of protein function molecular machines. They leverage nucleic acid aptamers for molecular recognition and intervention in the activity of target proteins, enabling the screening of high-specificity and high-affinity nucleic acid aptamers and the creation of molecular switches utilizing DNA structural and conformational changes. In comparison to protein-based tools, DNA molecules possess exceptional attributes such as ease of chemical synthesis, quality control, straightforward modification, biocompatibility, and low immunogenicity [97, 98]. Significantly, research in this domain has yielded effective tools for precise control of protein function at the cellular or organismal levels. For example, the Ren group [99] has reported the development of nucleic acid aptamers that block the function of IL-1α, exemplifying these advancements. Their work provides insights into the molecular structure and mechanisms underlying the intervention in IL-1α function.
Our group recently achieved a significant milestone by developing a DNA automaton designed to intricately modulate the interaction between TNF-α and its receptor TNFR-1. This automaton demonstrates remarkable selectivity, enabling the precise identification and regulation of programmed cell death in target cells within a multicellular environment [88] (Figure 3B). Additionally, it can provide activation components for immune receptors, showcasing its versatility and potential in immune-related applications. Notably, cytokine receptors share a common characteristic as single-pass transmembrane receptors, and they employ similar mechanisms involving ligand-responsive triggering to activate receptor oligomerization. In earlier research endeavors, our group pioneered a pioneering strategy for constructing DNA activators specifically designed for growth factor receptors. These activators were developed using DNA molecular dynamic self-assembly methods, which allowed us to achieve precise control over cell movement and in vivo tissue regeneration and repair. This control was facilitated through chemical (Figure 3C) or light-controlled receptor dimerization, ultimately leading to the activation of cell signal transduction pathways [100, 101] (Figure 3D). Moreover, Li et al. [102] described a reconfigurable two-dimensional DNA origami device with geometric patterns of CD95 ligands that can modulate immune cell signaling to alleviate rheumatoid arthritis. The device can reversibly transition between closed and open configurations in response to pH changes, displaying a hexagonal pattern of CD95 ligands that accurately mimic the spatial arrangement of CD95 receptor clusters on cell surfaces. This device allows precise spatial control of cellular signaling, expanding the understanding of ligand-receptor interactions and providing a promising platform for pharmacological interventions. Our team has recently harnessed DNA nanotechnology to design a non-heritable mechanosensor, allowing for customized mechanical signal transduction [103]. This modular platform can flexibly couple forces generated by various cells with specified receptor tyrosine kinase (RTK) signal output, possessing a fine-tuned force sensitivity and operating within the bio-relevant pN range. These technologies offer an innovative approach to precisely manipulate cell receptors through responsive receptor clustering to activate engineered components, presenting new possibilities for studying cellular signal transduction and biomedical applications. At present, analog peptides are increasingly concerned that, for example, peptide analogues of angiotensin as well as insulin peptide analogues for treating diabetic wounds has entered the clinical stage (Table 2).
Clinical application of various therapies to treat diabetic ulcers
| Type | Serial number | Clinical stage | Notes |
|---|---|---|---|
| Addressing therapy | NCT04098562 | Phase 2 | Antibacterial peptide LL-37 |
| NCT01143714 | Phase 4 | Collagenase | |
| NCT00316537 | Phase 1/2 | ATG002 is an agonist of the nicotinic acetylcholine (nACh) receptor pathway (pro-angiogenic drug) | |
| Cytotherapy | NCT06231771 | Phase 1/2 | Allogeneic mesenchymal stromal cells |
| NCT04104451 | Phase 1 | Umbilical cord endometrial MSCs | |
| Cytokine therapy | NCT01830348 | Phase 3 | A peptide analogue of angiotensin II |
| NCT06020664 | Phase 1 | Small molecule drugs | |
| NCT04323462 | Phase 4 | Insulin analog | |
| NCT06383013 | Phase 2 | Recombinant cytokines |
MSCs: mesenchymal stem cells
DNA nanotechnology offers a highly customizable, structurally predictable, and programmable approach to constructing molecular recognition modules, spatial scaffold modules, and dynamic assembly modules. Importantly, it allows for the precise response to specific environmental cues, such as ion strength, pH value, enzymes, and small molecules, which can trigger the restructuring or movement of DNA nanodevices. This versatility has paved the way for responsive receptor aggregation activation component engineering, a powerful tool for manipulating cell receptors under artificially defined conditions to achieve desired functions. Researchers have harnessed this technology to construct genetically engineered chimeric receptors with unique responsive activation characteristics. For instance, Wu’s group [104] developed DNA nanodevices responsive to various environmental inputs, including pH and ATP, by utilizing pH-responsive i-motif sequences and ATP-binding aptamers as responsive units. These devices selectively assembled receptors on the cell surface, such as MET and CD71, by constructing gates, thereby regulating HGF/MET signal transduction (Figure 3E). Recently, our group introduced a modular and programmable miRNA-responsive chimeric DNA receptor (miRNA-CDR). By combining miRNA responsiveness with chemically operated surface receptors, such as miR-122-CDR-MET, they could selectively sense and respond to elevated in situ levels of miR-122 after liver acute injury, leading to the activation of the c-MET receptor and accelerated hepatic cell proliferation, ultimately promoting liver regeneration [105] (Figure 3F).
The interaction between cell surface receptors and ligands is pivotal in regulating intracellular signal transduction. Most receptors do not act as monomers but require clustering to initiate downstream intracellular signaling pathways. Upon sensing extracellular stimuli, various cell surface receptors transition from freely diffusing monomers to dimers or higher-order oligomers. This transition triggers diverse downstream intracellular signaling pathways [106]. The aggregation of cell surface receptors is fundamentally a dynamic assembly process, and precise control of receptor assembly at the molecular level can effectively regulate intracellular signal transduction. For instance, our group has devised a receptor control strategy based on DNA origami, leading to the construction of DOTA. This technology enables the programmable arrangement of monovalent or bivalent ligands at the nanoscale, allowing precise programming of the oligomerization of RTK receptors. This achievement facilitates controlled chemical stoichiometry, valency, and cell behavior switching distances [94]. Furthermore, a unique DNA aptamer regulator, IR-a62, has been developed for the IR. This aptamer regulator selectively activates or inhibits IR activation by controlling its concentration, demonstrating the potential for precise control of receptor function [107].
Cytokines hold immense therapeutic potential, and extensive efforts have been directed towards harnessing them for the treatment of cancer and various diseases (Table 3). However, several challenges, including their short half-life, cytotoxicity, and off-target effects, have significantly limited their clinical utility. In recent years, researchers have embarked on multifaceted approaches to engineer cells, cytokines, and receptors, leading to the emergence of artificial cytokines that are now widely employed in drug development and disease treatment, particularly in diabetic wound healing. Although the application of artificial cytokines is currently less common, the principles of cytokine modification and cell engineering provide optimism for their future role in enhancing wound healing outcomes.
Applications of artificial factors for disease therapy
| Artificial factors | Characteristics | Disease | Clinical stage | Reference(s) |
|---|---|---|---|---|
| IFN-α mutant | Mutation | Cancer | Preclinical | [108] |
| Myeloid-biased, IL-10 variants | Mutation | Autoimmune diseases | Preclinical | [109] |
| PEG-IL-2, NKTR-214 | PEG modification | Solid tumors | Phase I/II | NCT02869295, NCT02983045 |
| IL-2-mutein-Fc, AMG592 | Mutation and fusion with Fc protein | Autoimmune diseases | Phase I | NCT03451422, NCT03410056NCT03422627 |
| Chimeric cytokines fused with the antibody | Type 2 diabetes | Phase I | [61] | |
| PCCPs | The pH-response properties were grafted onto the polypeptides | Cancer | Preclinical | [83] |
| HGF mimics | DNA aptamer | Fulminant hepatitis | Preclinical | [83] |
| IR-a62 | DNA aptamer modulator | diabetes | Preclinical | [107] |
IFN: interferon; IR: insulin receptor; HGF: hepatocyte growth factor; PCCPs: pH-controllable cell-penetrating polypeptides
Looking ahead, the principles of modifying natural cytokines and the techniques of cell modification hold great promise for advancing diabetic wound healing. Through the modification of crucial cytokines involved in the diabetic wound healing process, such as IL-1β, IL-2, TGF-β, PDGF, and FGF-2, by employing domain modification, PEGylation, lipidation, and other chemical modifications, along with fusion with structural domains like antibody fragments to enhance receptor affinity, prolong half-life, and mitigate toxic side effects, we anticipate significant progress in promoting diabetic wound healing. Additionally, the modification of corresponding cells to increase cytokine production offers another avenue to expedite the healing of diabetic wounds. Engineered cells and cytokines hold vast prospects in various medical fields. Particularly in disease treatment and regenerative medicine, they possess immense potential. Engineered cells can be manipulated to enhance their therapeutic effects by adjusting their functions and characteristics. They can replace or complement traditional drug treatments for various conditions such as cancer, cardiovascular diseases, and neurodegenerative disorders. Engineered cells and cytokines can also be utilized to cultivate artificial tissues and organs for the repair and regeneration of damaged tissues or organs. This technology offers new solutions for organ transplantation, tissue repair, and regenerative medicine, providing better treatment options for patients. With continuous technological advancements and in-depth research, it is believed that engineered cells and cytokines will play an increasingly crucial role in the future, bringing more hope and well-being to human health.
Recent years have witnessed the development of artificial factors based on peptide and nucleic acid classes. Peptides derived from cytokines and nucleic acid analogs have demonstrated the potential to replace natural cytokines, while peptide and nucleic acid-based agonists and antagonists have been designed to regulate the binding of cytokine-specific receptors, thereby influencing downstream signaling pathways. While most artificial factors based on peptides and nucleic acids are currently in the design phase, with limited clinical applications, their unique advantages hold the promise of future applications in diabetic wound healing and other disease treatments.
In conclusion, the field of cytokine engineering is poised for significant advancements, offering new avenues for improving therapeutic outcomes in diabetic wound healing and various other medical applications. The integration of artificial cytokines, cell modifications, and innovative peptide and nucleic acid-based approaches presents a comprehensive strategy to address existing challenges and pave the way for more effective wound healing treatments in the future.
bFGF: basic fibroblast growth factor
CAR: chimeric antigen receptor
ECM: extracellular matrix
IL-1β: interleukin-1β
IR: insulin receptor
MSCs: mesenchymal stem cells
TGF-β: transforming growth factor-β
TNF-α: tumor necrosis factor-α
VEGF: vascular endothelial growth factor
We would like to acknowledge the funding support from Joint Institute of Tobacco and Health, the National Natural Science Foundation of China, the Scientific Research Program of FuRong Laboratory, and the Natural Science Foundation of Hunan Province.
XZ: Conceptualization, Writing—original draft. HHW: Conceptualization, Writing—review & editing. TW, XF, TZ, RQ, and YW: Writing—original draft. MW, YZ, ML, NC, and YG: Writing—review & editing.
The authors declare that they have no conflicts of interest.
Not applicable.
Not applicable.
Not applicable.
Not applicable.
The study was supported by the Joint Institute of Tobacco and Health [2022539200340039]; the National Natural Science Foundation of China [22177030]; the Scientific Research Program of FuRong Laboratory [2023SK2104]; and the Natural Science Foundation of Hunan Province, China [2023JJ40814]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
© The Author(s) 2024.
Copyright: © The Author(s) 2024. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
