Abstract
The crown-like shaped viruses known as coronaviruses which were first reported in the 1960’s have caused three epidemics in the past two decades namely, coronavirus disease-19 (COVID-19), severe acute respiratory syndrome (SARS), and Middle East respiratory syndrome (MERS). SARS coronavirus 2 (SARS-CoV-2) was first reported in the latter half of December in Wuhan, a city of China, with people affected by deadly pneumonia with unknown etiology. Since then, the world has experienced two phases of virus spread with different symptoms and disease severity. This review embarks on the journey to investigate candidate molecules of this virus which can and are being investigated for various vaccine formulations and to discuss immunity developed against this virus.
Keywords
Severe acute respiratory syndrome coronavirus 2, Wuhan, vaccine, immunity, challengesIntroduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiological agent of coronavirus disease-19 (COVID-19) has created havoc worldwide, resulting in more than 3 million deaths in the past one and half years [1–3]. The surge in the number of cases in the pandemic can be brought under control with the development of effective vaccines. Several vaccines have been developed and are being developed based on limited knowledge of protective immunity and experience with SARS-CoV and the Middle East respiratory syndrome (MERS) [4]. Currently, there are now around 270 potential COVID-19 vaccines under development. The newly developed vaccines belong to various platforms which include RNA and DNA vaccines, live attenuated, subunits, etc. [5–13].
During SARS-CoV-2 infection, antibodies against spike (S) protein and nucleoprotein (N) are produced. On the basis of virus challenge research in animals, it was concluded that antibodies against S proteins are neutralizing in nature and are linked to protective immunity [14]. Studies have demonstrated that COVID-19 patients and people who have taken vaccinations have shown a reduced rate of SARS-CoV-2 infection [15–17].
Several vaccine candidates are efficacious and induce antibodies against S protein. These are known to have neutralizing activity against wild variants and currently circulating D614G variants [14].
The continuous evolution of this virus results in the changes that can affect immunity induced by the vaccine. These mutations affect the transmission and severity of the disease [4]. A brief overview of the immune response to SARS-CoV-2 is presented here, followed by insights into vaccine-induced immune protection.
Cell surface receptors
Angiotensin-converting enzyme 2
Angiotensin-converting enzyme 2 (ACE2) is a homolog of ACE that controls the blood pressure and electrolyte balance by stimulating the angiotensin II (Ang II) type I receptors in the rennin-angiotensin aldosterone system (RAAS). There are also increased fibrosis, inflammation, thrombosis, and pulmonary damage [18]. ACE2 converts Ang II, produced from Ang I to Ang 1–7. This Ang 1–7 activates the Mas receptor which is known to protect against lung injury [19, 20]. By interacting with ACE2, the S protein’s receptor binding motif reaches host cells. Endocytosis occurs in this complex and forms an endosome after SARS-CoV-2 activates the transmembrane serine protease 2 (TMPRSS2) or S priming. After that, the endosome is acidified, allowing the virus’s encapsulated single-stranded RNA to be released into the cytoplasm for reproduction and translation. There is a higher binding affinity of SARS-CoV-2 to these receptors. This could be the explanation for easier transmissibility. SARS-CoV-2 increases the nuclear factor kappa B (NF-kB) after infection. The pro-inflammatory cytokines interleukin-6 (IL-6) and tumor necrosis factor alpha (TNF-α) are released as a result [18]. A number of factors have been linked to changes in ACE2 expression as well as the severity and progression of COVID-19. The expression of ACE2 in the lungs and SARS-CoV-2 viral load increase with age. This is the potential explanation for the increased risk of contracting severe COVID-19 in older adults (aged > 60 years). Male predominance is also observed among COVID-19 patients which could be due to gender disparities in ACE expression on chromosomes, sex hormone-driven immune system control, or RAAS [19, 20].
Glucose-regulated protein 78
Also known as heat shock protein A5 or binding immunoglobulin protein is a chaperone protein, released from the endoplasmic reticulum. Under normal conditions, glucose-regulated protein 78 (GRP78) is located in the lumen of the endoplasmic reticulum and results in inhibition of protein synthesis, increased protein folding and cell death after binding to inactivating enzymes. GRP78 is a potential getaway for entry of virus in COVID-19. It controls the release of unfolded protein response under cellular stress conditions, which leads to the entry of SARS-CoV-2. Once GRP78 is translocated to the plasma membrane, it further facilitates the entry of viruses via motifs on S protein and molecular docking [21, 22]. SARS-CoV-2 binds to GRP78 via a molecular docking mechanism that occurs when the GRP78 substrate-binding domain area engages with SARS-CoV-2 [23].
COVID-19 patients have increased gene expression for GRP78 and there is increased release of GRP78 from damaged airway cells, which results in severe lung injury and trauma during the infection. This may be responsible for augmented inflammation in COVID-19 patients. Thus, GRP78 mediates binding to the S protein of SARS-CoV-2 and thus may represent a potential target for therapeutic treatment for COVID-19 [18]. According to studies, the celecoxib derivative AR12 (OSU-03012) can be used as a treatment alternative to reduce protein renaturation and S protein synthesis by inhibiting GRP78 ATPase activity with the celecoxib derivative AR12 (OSU-03012) [24, 25].
Transmembrane serine protease 2
It’s a protease produced by the host cells that primes the S protein, allowing binding to ACE2. The important cellular surface receptor mediating the SARS-CoV-2 entry is ACE2. Once the S protein of SARS-CoV-2 binds to ACE2, it undergoes conformational changes followed by proteolytic degradation by host cell proteases, notably TMPRSS2 [26]. Thus, it increases infectivity and transmission of the virus. In addition to cathepsin B/L mediated endocytosis, TMPRSS2 cleaves the S protein and also activates the entry of SARS-CoV into host cells [27]. As the virus binds to ACE2, researchers discovered that fusing of plasma and viral membrane during the entrance mechanism causes TMPRSS2 to trigger extracellular priming of S protein [28]. The expression of TMPRSS2 is required for viral dissemination and pathogenicity. This gene’s genetic variation may influence the host’s predisposition to infection and virus clearance. The male gender is more susceptible to infection because the expression of this receptor is much more in androgen sensitive organs including the prostate and testis. This could be the plausible reason for the difference in infection rates between men and women [28].
Ezrin (receptors)
Ezrin (EZR) is a protein encoded by the EZR gene and belongs to the family EZR-radixin moesin. The role of EZR has been described in the transmission of human immunodeficiency virus (HIV) but in SARS, it causes decreased entry of the virus by interacting with the SARS-CoV S protein [29], causing reduced viral entry. This can be considered as a potential therapeutic alternative to prevent SARS-CoV-2 infection. As EZR inhibits ACE2 and the Toll-like receptors (TLRs), the crucial receptors participating in the pathogenesis of COVID-19, an EZR agonist or molecule can be considered as a strategic therapeutic approach to affect SARS-CoV-2 viral entry [18]. It suppresses the inflammation seen in viral pneumonia [29], a significant pathophysiological consequence seen in COVID-19. As a result, more research is needed to fully comprehend its role in the SARS-CoV-2 virus infection.
TLRs
TLRs belong to innate immune receptors that activate the innate immunity and regulate cytokine expression [30]. TLRs have 10 members in the human family. TLRs are present in the cell membrane, and endosomes are their other location for receptors like TLR3, TLR7, TLR8, and TLR9. TLRs are expressed by a variety of immune cells, including dendritic cells (DCs), macrophages, natural killer (NK) cells, and cells of adaptive immunity, such as T cells and B cells [31, 32]. TLR3 detects double-strand RNA (dsRNA), TLR4 detects lipopolysaccharide (LPS), TLR7/8 detects single-strand RNA (ssRNA), and TLR9 detects unmethylated cytosine-guanine (CpG) DNA [31]. TLR signals are transduced by two main pathways: myeloid differentiation primary response 88 (MyD88) and Toll/interleukin-1 receptor/resistance protein (TIR) domain-containing adaptor-inducing interferon (IFN) [TRIF, also known as Toll-IL-1R-containing adaptor molecule 1 (TICAM)]. TNF receptor associated factor (TRAF) and interleukin 1 receptor associated kinase (IRAK) are signaling proteins that activate NF-kB and interferon regulatory factor (IRF), resulting in the production of type I IFN and proinflammatory cytokines like IL-1, IL-6, TNF, and IL-12 [30].
TLRs in SARS-CoV-2
TLR activation aids in the eradication of viruses, but it can also harm the host due to prolonged inflammation and tissue death. Once SARS-CoV-2 activates TLRs causes the inflammasome activation and IL-1β production which activates IL-6. This may result in a bad prognosis in COVID-19 patients. TLRs have a dual role in viral infections activating Janus kinase/signal transducers and activators of transcription (JAK/STAT) [30]. In COVID-19 patients, there is lymphopenia and elevated serum levels of pro-inflammatory cytokines, such as TNF-α, IL-6, and IL-2 receptor (IL-2R). These changes are linked to the severity of the disease. TLR activation in COVID-19 infection, according to Conti et al. [33], could result in the generation of proinflammatory cytokines such as IL-1β. Other studies by Totura et al. [34] suggested that TLRs activate both TRIF and MyD88 signaling pathways, resulting in the most effective antiviral defense against SARS-CoV and deadly SARS-CoV illness if TLR signaling pathways are disabled.
TLR7/8 in COVID-19
TLR7/8 are genes on the X chromosome that detect ssRNA such as SARS-CoV-2 genome. TLR7 could detect when superficial S glycoprotein binds on the envelope of the virus to ACE2. TLR7 activation results in increased synthesis of IL-1, IL-6, monocyte chemoattractant protein 1, macrophage inflammatory protein (MIP)1α, TNF, and type I IFN [35]. Increased TLR7 expression leads to a better prognosis in ssRNA viral infections, resulting in a stronger immune response. SARS-CoV-2 has more ssRNA motifs that could interact with TLR7. This has been observed in the results of whole-genome sequencing of SARS-CoV-1, MERS-CoV, and SARS-CoV-2 revealed that TLR7 could be more involved in the pathogenesis of SARS-CoV-2 as compared to SARS-CoV and MERS-CoV [36]. TLRs once activated result in the release of pro-inflammatory cytokines like interleukins: IL-1, IL-2, IL-6, IL-8, IL-12 as well as IFN-α.
TLR2/6/9 agonists also activate innate immune cells and lung epithelial cells, causing them to generate anti-infection factors. Several clinical trials are being undertaken to evaluate the role of anti-inflammatory factors in the death caused by lung injury in COVID-19 patients.
Immune response to SARS-CoV-2
Understanding the molecular and cellular mechanisms underlying the immune response elicited by SARS-CoV-2 can help researchers better understand factors that could affect the epidemic’s outcome, such as whether people develop lasting immunity after recovery from infection and the best vaccine-induced immunity to provide adequate protection, thus help to develop therapeutic strategies and vaccines [37].
Protective immunity to a pathogen’s correlates can be thought of as quantitative indications that reliably identify individuals as immune to specific outcomes like infection, illness result, or transmission risk. The adaptive immune response gives the strongest long-term protection after infection with SARS-CoV-2 or immunization [38].
Innate immunity, a less specific component, is the initial line of defense against all infections, including SARS-CoV-2. The innate immune system recognizes the characteristics of a pathogen and then responds with inflammation, phagocytosis, cytokine release, stimulation of type I IFN activation of NK cells, and/or a complement system. When the innate immune system fails to clear an infection, the adaptive immune system is notified and activated [38, 39].
Innate immunity includes the majority of the body’s defense mechanisms and is present before the onset of infection. When a virus enters the body, cells can recognize markers present on the virus resulting in non-specific antiviral activity and cells of the innate system (such as macrophages, neutrophils, DCs, and others) are activated to remove pathogens and foreign cells from the body. Invasion of monocytes/macrophages, neutrophils, and other adaptive immune cells results in an increase in pro-inflammatory cytokines. Thereafter, the innate response activates the adaptive immune response.
T cells (cellular response) and B cells (adaptive immune response) are the two types of white blood cells that make up the adaptive immune response (antibody response). Various types of antibodies are produced by memory B cells to neutralize the virus or virus-infected cells, while memory T cells stimulate antibody synthesis when virus-infected cells are killed. Despite the fact that both T cell and memory B cell immune responses have been induced in both SARS-CoV-2 infected and vaccinated people, clear correlates for protective immunity have yet to be identified [40, 41]. We herein have tried to explain the immune response, against SARS-CoV-2.
Innate immune response
The SARS-CoV-2 virus enters the body via the naso-oral pathway and infects cells in the lungs that express the ACE2 receptor, such as pulmonary epithelial cells and type II alveolar cells. After the virus penetrates the target cell, the host immune system identifies the complete virus or its surface epitopes, prompting an innate or adaptive immune response [42].
Following COVID-19 infection, there is a robust innate immune response, as evidenced by increased levels of C-reactive protein (CRP) and serum amyloid A (SAA) [43]. Innate immune cells such as macrophages, monocytes, and DCs become activated after recognizing damage-associated molecular patterns (DAMPs) are pathogen-associated molecular patterns (PAMPs) through their pattern recognition receptors (PRRs) [44]. PRRs on immune cells, primarily TLRs 3, 7, and 8, Nod-like receptors (NLRs), and retinoic acid-inducible gene (RIG)-like receptors (RLRs), are the first to recognize the virus, resulting in increased activation of various signaling pathways and transcription factors, as well as IFN production. The expression of cytokines, chemokines, and adhesion molecules is triggered when PRRs bind to PAMPs.
The generation of antiviral immunity in the lungs is controlled by nucleic acid detecting receptors and subsequent intracellular signaling pathways. Endosomal receptors (TLR3, TLR7, TLR8) and cytosolic RNA sensors [melanoma differentiation-associated gene 5 (MDA5) and RIG-I] detect viral RNA, triggering a signaling cascade that activates several transcription factors (such as neurofeedback (NFB) IRF3, IRF7, and others). IFN receptor signaling activates interferon-stimulated genes (ISGs) in both virally infected and adjacent cells via JAK/STAT pathways, increasing the innate immune response [45].
The nuclear factor-B (NF-B) and IRF3/7 signaling pathways are activated, resulting in increased production of pro-inflammatory cytokines such as TNF, IL-1, IL-6, IL-17, and IFN. These cytokines are well-known for their importance in virus clearance. There are increased levels of interferon gamma-inducible protein 10 (IP-10) fibroblast growth factors 2 (FGF2), granulocyte-colony stimulating factor (GCSF), granulocyte-macrophage colony-stimulating factor (GMCSF), monocytes chemoattractant protein-1 (MCP-1), MIP-1, TNF-α, platelet-derived growth factor subunit B (PDGFB) [46].
Certain cytokines, such as IL-6, IL-7, IL-1, IL-10, IP-10, and TNF determine the severity of the infection and contribute to a severe inflammatory response known as cytokine storm, which leads to severe conditions such as acute respiratory distress syndrome (ARDS), disseminated intravascular coagulation, or multiple organ failure. C-X-C motif chemokine ligand 8 (CXCL8), CXCL1, CXCL2, CXCL10, C-C motif chemokine ligand 2 (CCL2), and CCL7 are involved for neutrophil recruitment, and CXCL6, CXCL11, CXCL2, CCL3, CCL4, CCL7, CCL8, and CCL20 for monocyte and other immune cell recruitment [47, 48].
Along with the abovementioned cytokine storm, the severity of COVID-19 illness also depends on dysregulated or excessive immune response and macrophage activation syndrome.
Role of IFNs
SARS-CoV-2 induces diminished or delayed type I IFN responses, thus generating a replication permissive microenvironment early in the disease course [49]. SARS-CoV-2 replicates to larger titers in the absence of efficient IFN expression, resulting in an exacerbated inflammatory response. This explains the large viral loads reported in COVID-19 patients at the onset of disease symptoms, the exceptionally extended viral incubation time, and the high prevalence of asymptomatic or pauci-symptomatic patients [50].
IFNs function by instructing uninfected cells to degrade RNA and limit protein synthesis. type I and type III IFN-responses are the first innate antiviral actions, occurring within hours of infection, and operate as barriers to virus reproduction and spread [51]. According to current findings, some patients with severe COVID-19 have a delayed or absent induction of type I and III IFN [52]. Nsp 13, Nsp15, and open reading frame 9b (ORF9b) target the IFN pathway, while Nsp13 and ORF9c target the NF-kB pathway.
Type I IFNs, through diverse mechanisms, can directly block virus multiplication and improve both innate and adaptive immunity. Various mechanisms include IFN-inducible dsRNA-dependent protein kinase R (PKR), myxovirus resistance protein 1 (MxA/Mx1) GTPase polymerization, 2’-5’-oligoadenylate synthetase (OAS), IFN-induced transmembrane proteins (IFITMs), and the tripartite motif (TRIM) family of molecules.
Adaptive cellular and humoral immunity
The adaptive immune system, also known as the acquired immune system responds to infections in an antigen-specific manner. Cellular immunity, which is mediated by T cells, and humoral immunity are the two principal arms of this system. SARS-CoV-2 infection activates macrophages and causes them to secrete inflammatory cytokines, which causes T and B cells to differentiate. Multiple T cell subsets [such as T-helper 1 (Th1) and Th17] are activated, releasing cytokines that help to amp up the immunological response. Stimulation of Th1/Th17 cells with viral epitopes in the helper T cell subset may lead to aggravated inflammatory responses resulting in “cytokine storms” that lead to consequences like pulmonary edema and pneumonia [48].
Lympopenia, increased neutrophil-to-lymphocyte ratio (NLR) cytokine release syndrome (CRS), lymphocyte exhaustion and dysfunction, antibody-dependent enhancement (ADE), and abnormalities of monocytes and granulocytes are the major events implicated in the dysregulation of immune response and the immunopathogenesis of COVID-19 in these patients. In severe cases, T cell subsets such as CD4+, CD8+, and memory T cells are reduced, reflecting an insufficient antiviral response following SARS-CoV-2 infection [43].
Cytotoxic T cells recruited to the site of infection try to kill virus-infected cells in the lungs and are hyper-activated and exhausted whilst CD4+ T cells have decreased inflammatory response during SARS-CoV-2 infection. SARS-CoV-2 is unable to multiply within T cells, resulting in fruitless infection, cell death, protracted virus clearance, and a weakened adaptive immune response. B cells/plasma cells identify viral proteins and create SARS-CoV-2-specific antibodies, which could give systemic immunity [53]. Other immune cells such as NK cells, memory and regulatory T cells (Tregs), and B cells are lost in severe immunological disorders [53].
The humoral response to SARS-CoV-2 was discovered to be similar to that of other coronavirus infections, involving the generation of immunoglobulin G (IgG) and IgM antibodies. At the outset of SARS-CoV-2 infection, B cells elicit an early response against the N but antibodies against the S protein were found 4–8 days after the onset of symptoms. The absence of glycosylation sites on N results in the production of N-specific neutralizing antibodies at an early stage of acute infection is highly immunogenic. Studies have shown that SARS-CoV-specific IgA, IgG, and IgM antibodies were detected after the onset of symptoms. IgG levels persist for a longer time; however, IgM levels began to fall after 3 months [53].
Antibodies against the S1 glycoprotein, particularly its receptor attaching domain, neutralize SARS-CoV-2 by blocking the virus from binding to ACE2 and thereby severely limiting virus cell entrance. The existence of neutralizing antibodies against SARS-CoV-2 is the strongest current indicator of protection against reinfection or breakthrough infection in vaccinated people [54].
Age-related differences in the immune response to SARS-CoV-2
Age itself is associated with an increase in the systemic and tissue levels of different pro-inflammatory mediators and reactive species of oxygen, in realizing a condition defined as “inflame-aging” [55].
COVID-19 causes more severe symptoms in older persons, however, it also affects younger people. It has been observed that young people and children shed equivalent levels of SARS-CoV-2 virus to older adults during infection [56]. This could mean that younger people can tolerate higher viral loads better, or that the immune response to infection in older people has a bigger factor in severity and symptoms. This disparity in symptoms between age groups could be due to younger people’s decreased expression of the SARS-CoV-2 cell entry receptor, ACE2. ACE2 is a protein on human cell surfaces through which SARS-CoV-2 gains entry into the cells. The predominant cell type infected by SARS-CoV-2 is respiratory epithelial cells, and the intrinsic antiviral responses of these cells alter with age. The senescence of respiratory epithelial cells as people get older may help to sustain viral infection and local tissue inflammation [54, 55].
Children develop symptoms that may be milder due to more active innate immune responses, healthier respiratory tracts due to less exposure to air pollution or cigarette smoke, and fewer comorbidities than adults [57]. IFN, which inhibits viral reproduction, and macrophages, which engulf and digest viruses, are released as part of the innate immune response. In addition, children express fewer ACE2 receptors in their airway cells than adults.
DCs (both myeloid and plasmacytoid subsets) in the periphery exhibit a considerable reduction with age. Because DCs are the most essential antigen-presenting cells (APCs) and serve as a link between innate and adaptive immunity, their loss over time contributes significantly to the increased susceptibility to infections as people get older [56].
Immune evasion by SARS-CoV-2
The prevalence of a specific viral strain, as well as its fitness and capacity to evade existing immunity, is determined by a number of factors. Virus transmissibility and replication competence are the most important parameters that drive fitness, but numerous other factors, such as viral receptor affinity, can also play a role. Immune evasion, on the other hand, is usually driven by a reduction in neutralizing antibody recognition, with serotype creation being the most extreme form of this process [58]. Changes in viral surface proteins involved in cell attachment and/or fusion are thought to be the main driver of increased transmissibility and/or virulence. Due to unrestricted virus reproduction, these viruses decrease antiviral IFN responses by evading innate immune cells.
SARS-CoV-2 evasion of viral RNA recognition
SARS-CoV-2 adapts to the host and evades the immune system, causing changes in its genome. Amino acid substitutions that cause major changes in the structure and stability of epitopes in the REM sleep behavior disorder (RBD) may be more significant, as these mutations may result in lower antibody-target protein binding affinity and energy. As a result, antibodies that disrupt key epitopes in the RBD are critical in degrading the RBD’s ACE2 receptor binding [59].
As SARS-CoV-2 replicates in humans under selective pressure from natural and vaccine-induced immunity, variants of concern (VOCs) with greater transmissibility or virulence continue to arise [60].
The newly discovered variant B.1.617 is divided into three sublineages (B.1.617.1, B.1.617.2, and B.1.617.3), each with its own mutational profile. Only sublineage B.1.617.2 or Delta is currently recognized as VOC on a global scale. S mutations T19R, G142D, 157–158, L452R, T478K, D614G, P681R, and D950N characterize it. The S protein of the B.1.351 virus largely mimics the structure of the G614 trimer, with nearly comparable biochemical stability. The RBD has not changed significantly as a result of N501Y, K417N, or E484K, however, the lack of salt bridges between Lys417 and ACE2 Asp30 and Glu484 and ACE2 Lys31 mitigates N501Y’s greater receptor affinity [58, 59, 61].
SARS-CoV-2 evasion of RLR signaling and IFN production
The structural proteins [membrane (M) protein and N] or Nsps (Nsp1, Nsp3b and Nsp6) or PLpro are used by SARS-CoV-2 and other coronaviruses to suppress the signaling cascade (papain-like protease). The JAK/STAT pathway is inhibited by these viral accessory proteins, resulting in gene suppression by IFN-stimulated response element (ISRE) promoters. SARS-nucleocapsid CoV’s protein (N) interferes with IRF3’s function. RNA binding activity at the first recognition stage of viral RNA may contribute to immune evasion, even though it does not form a complex with RIG-I or MDA5. On the one hand, the production of pro-inflammatory cytokines and type I IFNs creates an antiviral immune microenvironment, controlling viral synthesis and infection; however, the SARS-CoV-2 shuts down these signaling pathways to counteract the immune response via various strategies, such as leukopenia [62]. The immunopathogenesis of COVID-19 has been summarized in Table 1.
| COVID-19 immunopathobiology | Outcome of immunopathogenesis |
|---|---|
| Lymphopenia | Decrease in Th cells, cytotoxic cells, memory T cells, Treg cells, B cells, NK cells |
| Neutrophilia | Increased neutrophil: lymphocyte ratio |
| Granulocyte & monocyte abnormalities | Decrease in eosinophils, basophils, monocytes |
| Cytokine release syndrome | Increased production of pro-inflammatory cytokines: IL-6, IL-12, IL-18, IL-17, IL-33. IL-1β, TNF-α, IFN-α, IFN-γIncreased production of chemokines: CCL2, CCL3, CCL5, CXCL8, CXCL9, CXCL10 |
| Lymphocyte exhaustion | T cell and NK cell exhaustion |
| Antibody dependent enhancement | Increased viral entry into host cells |
Studies have shown that due to the reduced synthesis of type I and III IFNs with sufficient ISG expression, as well as increased chemokine secretion, SARS-CoV-2 infection causes an overall decrease in antiviral gene transcription. COVID-19 severity could be caused by a decrease in the innate antiviral response, as well as hyper-inflammation. SARS-CoV-2 infection decreases the immune response against the virus by increasing the fatigue of effector T cells, in addition to reducing T cells (Table 2) [46].
| Immunologic changes | COVID-19 |
|---|---|
| T cell responses | Lymphopenia in severe cases. |
| CD8+ cells | Severe lymphopenia, a predictor of severe COVID disease. |
| Th1-Th2 responses | Severe systemic inflammatory response accompanied by cytokine response.Plays a part in the activation of inflammasomes. |
| Eosinophils | The number of circulating eosinophils has decreased. |
| Cytokine storm | In severe disease, large numbers of innate and adaptive cytokines are released. |
| Acute phase reactants | Initial high values are predictors of severe disease. |
| Specific antibody levels | Virus-specific IgM rises during the acute phase, followed by virus-specific IgG during the recovery phase. |
After RLR activation, TANK-binding kinase 1 (TBK1) is recruited to the RLR/mitochondrial antiviral signaling protein (MAVS) signalosome to activate the transcription factors (TFs), IRF3, and NF-B, which activate IFN and IFN gene expression. By altering this signaling complex, a few SARS-CoV-2 proteins have been identified that antagonize dsRNA recognition. In HEK293T cells, Nsp6 and Nsp13 have been discovered to decrease IFN promoter reporter activity in response to RIG-I caspase recruitment domain family member (CARD). This could be due to a decrease in IRF3 (S396) phosphorylation in Nsp6 or Nsp13 expressing cells after pI:C transfection. Few studies have found Nsp13-TBK1 interactions, but none have found Nsp6-TBK161 interactions or Nsp6 suppression of IFN induction. These inconsistencies could be explained by differences in Nsp6 expression levels between experiments, implying that protein expression levels during infection are crucial for immunological antagonism [62].
Activated NF-kB works as a transcriptional activator for a variety of pro-inflammatory cytokines with an NF-kB-response element after translocation into the nucleus. After being phosphorylated by ubiquitin kinases, the IRF3 homodimerizes and travels inside the nucleus, activating type I IFN transcription. Type I IFNs activate the JAK/STAT signaling pathway through the IFN a/b receptor (IFNAR), which is followed by the cytoplasmic protein JAK1 and tyrosine kinase 2 (TYK2) kinases phosphorylating STAT1 and STAT2. STAT1 and STAT2 heterodimers translocate into the nucleus and recruit ISRE transcription to their promoter.
COVID-19 vaccines
Publication of the genome sequence of a COVID-19 virus on January 11, 2020, an effort of unparalleled speed and magnitude set out to develop a vaccine for this contagious disease. Figure 1 depicts a complete SARS-CoV-2 vaccine that should meet the main requirements like (1) protection not only from severe disease but also impart prevention in vaccinated groups, these may include the immunocompromised peoples, (2) impart long term immune memory, (3) the ability of the companies producing the vaccine should be high so that every individual in this world can have vaccination shots annually and for the booster doses as well [66].
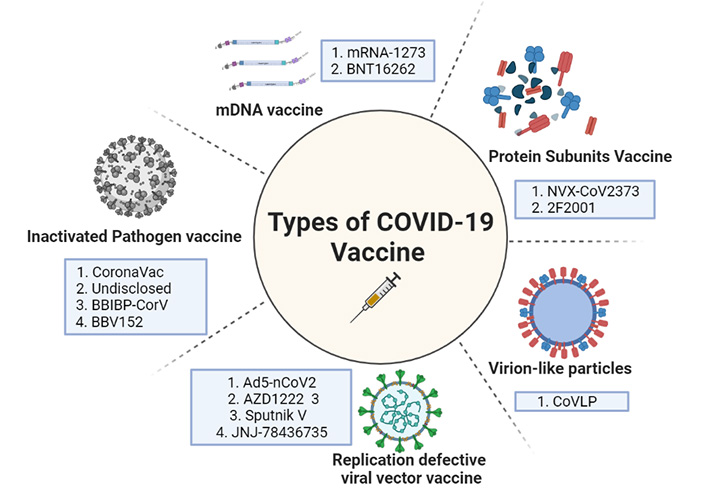
The diagrammatical representation showing the different types of vaccines against COVID-19. mRNA: messenger RNA
The vaccination story comes from Edward Jenner in the year 1796, on May 14th, where he did an experimental inoculation of a young boy with pus acquired from a cowpox-infected milkmaid, which led to his smallpox vaccine. This chance observation has revolutionized the field of medicine. Vaccination and vaccine discovery has come a long way since then! Traditional vaccines have been there for several pathogens and are efficient enough, but slowly evolving technologies like recombinant DNA technology (RDT) give space to more complex techniques involving combining newer and more than one option in vaccine treatment strategies [67].
Attenuated SARS-CoV-2 viral vaccines
Attenuated vaccines are based on a living microbe weakened not to cause disease. Microbes that have been attenuated retain their ability to multiply in vivo, resulting in restricted illness and they effectively stimulate the immune system and induce an intense and persistent immune memory that is efficacious in preventing infection. The YF-S0 vaccine, which uses a live-attenuated yellow fever 17D (YF17D) vaccine as a vector to express a non-cleavable pre-fusion form of the SARS-CoV-2 S antigen is an example of an attenuated vaccination for SARS-CoV-2 [68].
mRNA vaccines
Modern therapeutics, based in Boston and the National Institute of Allergy and Infectious Diseases, collaborated to develop mRNA-1273. This is one of the first vaccine candidates which entered the clinical trials in just 63 days after the genome of SARS-CoV-2 was sequenced. A mRNA molecule in this vaccine design is capable of synthesizing a lipid nanoparticle (LNP) encapsulated pre-fusion version of the SARS-CoV-2 S protein. This form of the LNP encapsulated S protein makes its uptake easier by the host cells. The second candidate is mRNA-BNT162b2. Pfizer created the BNT162b2 mRNA platform in conjunction with BioNTech in Germany and Fosun Pharma in Shangai, China. It promotes the synthesis of SARS-CoV-2 S protein. A higher dose of RNA is required for mRNA than for delf-reoplicating RNA, which amplifies itself [69].
Vaccines with viral vectors that are replication-defective
CanSino Biologics developed the Ad5-nCoV vaccine (a China-based company), using human adenovirus serotype five vectors to deliver the message that codes for COVID-19 virus full-length S protein into human cells. An HRB26M mouse-adapted SARS-CoV-2 virus was used to challenge the immune mice. Immunized mice showed no signs of viral replication in their lungs or any other obvious viral symptoms. Immunized ferrets were subjected to wild-type SARS-CoV-2, and the results of vaccination immunogenicity and viral challenge protection were repeated [70]. Oxford University’s AZD1222 is a viral vectored vaccine, while AstraZeneca is another contender. This vaccine from Oxford University was the first to enter clinical trials using a weakened chimp adenovirus (ChAdOx1) platform. Because very few humans have had previous interaction with a simian virus, this was done to avoid the issue of pre-existing immunity to the vector. Another, Russian Research Institute, Gamelaya produced Sputnik V, which is the only heterologous prime-boost coronavirus vaccine. This vaccine candidate can circumvent the challenge of reduced immunogenicity due to immunoglobulins built against the viral vector after the initial immunization [71]. Here the adenoviral vector serotype used for the prime vaccination is more diverse from the adenoviral serotype used as a booster, thus giving an imperative and stronger immunity in the first shot itself. In the list of vaccine candidates, Janssen Pharmaceuticals developed JNJ-78436735. This comes under Johnson & Johnson Pharmaceuticals’ vaccine development division. This vaccine is made up of a replication-defective adenovirus 26-based vector that expresses the COVID-19 virus’s stabilized pre-fusion S protein, which was developed by Beth Israel Deaconess Medical Center researchers in Boston a decade ago [71].
Protein subunit vaccines
A Maryland based pharma company developed an inactivated, protein subunit vaccine—NVX-CoV2373. It comprises a pre-fusion full-length recombinant COVID-19 glycoprotein nanoparticle that was exposed in a baculovirus/Sf9 system and treated with Matrix M1 adjuvant. This saponin-based Matrix M1 adjuvant-based RBD-dimeric antigen, developed by Anhui Zhifei Longcom Biopharmaceutical and the Chinese Academy of Medical Sciences’ Institute of Microbiology, investigates the many forms of the whole S protein or its RBD. As a result, when these moieties engage with the ACE2 receptor on the host cell, the pre-fusion conformation changes to a highly stable post-fusion conformation. This strengthens the viral-host cell bond. These pre-fused viral proteins are more immunogenic, thus are explored well as lucrative vaccine candidates. This ZF2001 is the most recent subunit vaccine candidate in phase 3 clinical trials that was launched in December 2020 [69, 70].
Virus-like particle vaccine
Virus like vaccines [virus-like particles (VLP’s)] is basically made of empty virus particles lacking genetic material. These VLP’s like CoVLP represent many numbers of copies of the same antigen on their surface making it more immunogenic, thus helping to trigger an efficient immune response. It allows rapid uptake by APCs followed by phagocytosis and presentation by the DCs [71]. They are much more efficient than attenuated vaccines but because of their complexity in development and assembly, makes their production technically challenging. The first trials were done by a Canadian Pharmaceutical firm Medicago Inc. 180 participants were enrolled in the first phase of clinical trials (age 18–55). Adjuvants like CpG1018/As03 (DynaVac) helped in the induction of robust cellular and humoral immune response [72, 73]. Study results on CoVLP indicated that administration was well tolerated with only mild to moderate transient adverse events [74].
Inactivated pathogenic vaccine
The coronaVac vaccine was initially launched under PiCoVacc, which is a purified, inactivated viral vaccine with alum-adjuvant. This potential vaccine was created by propiolactone-activation of the CN2 strain of SARS-CoV-2 using a broncho-alveolar lavage of a hospitalized SARS-CoV-2 patient [71]. Sinopharm collaborated with Beijing Biological Products to create BBIBP-CorV, the second inactivated viral vaccine candidate. BBIBP-CorV was created by inactivating the 19nCoV-CDC-Tan-HB02 strain SARS-CoV-2 in Vero cells using propiolactone [72]. Adjuvant, aluminum hydroxide in this vaccine stimulates the receptor subunit Nod-like receptor family pyrin domain containing 3 (NLRP3) from the inflammasome and inflammasome-derived IL-1β and IL-18 release is observed at a marked increased level. These cytokines thus stimulate the proinflammatory cells of the immune system [73]. Bharat Biotech an India-based pharmaceutical company that produced Covaxin/BBV15 is a whole virion inactivated vaccine candidate approved by the Indian Council of Medical Research. This vaccine was created utilizing a new coronavirus that was identified by the Indian National Institute of Virology and propagated on Vero CCL81 cells before being inactivated with propiolactone.
DNA vaccine
The DNA-based platforms offer great versatility while handling the coded antigen and great potential to advance. Till date no single DNA vaccine has been registered for human use; however, they are commonly used in veterinary medication. Certain vaccines are stable and can quickly be produced in large quantities in bacteria. Once inserted by the myocyte pathway, DNA plasmids penetrate human cells, and a small local electrical pulse may boost their ability to do so (electroporation) [75]. Once entered by the APC pathway (Figure 2), plasmid DNA causes the cell to produce the target protein briefly. This way, DNA vaccination stimulates the APC and enhances the production of antibodies. A similar scenario can also be seen in gut-associated lymphoid tissues (GALTs) such as the Peyer’s patches [76].
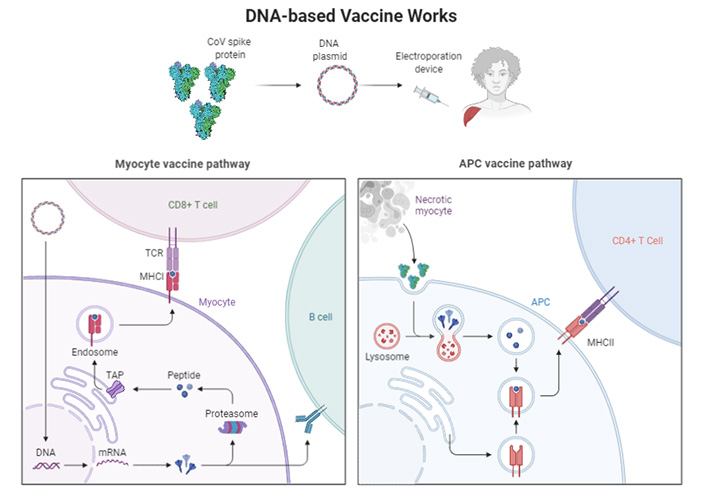
Activation of cellular pathways by the DNA based vaccines. TCR: T-cell receptor; MHCI: major histocompatibility complex I; TAP: transporter associated with antigen processing
The till date vaccine candidates available or which are under different phases of development are summarized in Table 3.
Advanced SARS-CoV-2 vaccine candidates
| Vaccine name | Clinical trial number | Manufacture | Phase trial | Number of participants | Immunity response | Efficacy | Ref. |
|---|---|---|---|---|---|---|---|
| mRNA-1273 | NCT04470427 | MODERNA/NIAID | 3 | 30,000 | CD4+ T cell activation reported Th1 skewed phenotype | 94.10% | [71] |
| BNT 162b2 | NCT04368728 | BioNTech/Pfizer | 3 | 44,000 | Virus specific Th1 and CD8+ T cell responses reported | 95% | [71] |
| Ad5-nCoV | NCT04526990 | CanSino Biological | 3 | 40,000 | T cell responses were observed in 88% of the participants | - | [71, 72] |
| AZD1222 | NCT04540393 | AstraZeneca | 3 | 30,000 | T cell responses observed in all participants | 62.10% | [71, 73] |
| Sputnik V | NCT04530396 | Gamaleya Research Institute | 3 | 40,000 | CD4+ and CD8+ T cell responses were observed in all participants | - | [71] |
| JNJ-78436735 | NCT04505722 | Janssen Pharmaceutical | 3 | 90,000 | CR4+ T cell responses in 80% of the participants | - | [71, 75] |
| 53coronaVac | NCT04456596 | Sinovac Research & Development Co. | 3 | 8,870 | Not reported | - | [71, 73] |
| BBIBP-CorV | NCT04560881 | Beijing Institute of Biotechnology | 3 | 63,000 | Not reported | 79.38% | [71, 74] |
| BBV152 | CTRI/2020/11/028976 | Bharat Biotech | 3 | 26,000 | Virus specific CD4+ and CD8+ T cell responses reported | - | [71, 73] |
| NVX-CoV2373 | NCT04611802 | Novavax | 3 | 45,000 | CD4+ T cell activation in all tested participants | - | [71, 75] |
| CoVLP | NCT04636697 | Medicago | 3 | 30,612 | Not reported | - | [71, 76] |
Ref.: reference. “-” in the efficacy column mentioned in Table 3 refers to the lack of information/data on that specific vaccine. Information on these clinical trials can be retrieved from https://clinicaltrials.gov/ct2/home
Development of SARS-CoV-2 vaccine
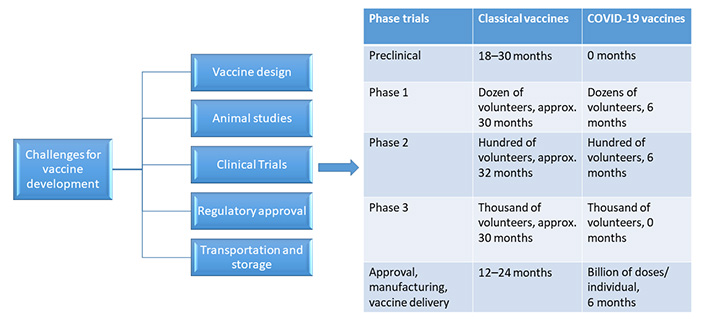
As history has shown, producing a vaccine is a lengthy process that takes anywhere between 10 and 15 years. The vaccine which was developed in the shortest of time was mumps, which took approximately 5 years. Therefore, it has been a challenge for developing the SARS-CoV-2 vaccine in a span of 12–24 months [75].
An exploratory stage is the first stage of vaccine development where hundreds and thousands of natural and synthetic antigens that could be exploited to develop a vaccine are explored by computational modeling for basic laboratory bench research. It could aid in the prevention or treatment of an illness. The suitably investigated antigens are next examined for immunogenicity in cell-culture or tissue-culture systems, followed by testing on animal models to assess the candidate vaccine’s safety. It is said as ‘valley of death’ when any vaccine candidate moves from the lab to clinical studies, where the majority of molecules fail. After the vaccine candidate clears these stages with proven immunogenicity and efficacy, progress is made to human clinical trials. Another important concern during the testing of vaccines in clinical trials is that usually it is being done in small population groups and then fast-tracked to larger population sizes. As vaccine production scales up, there’s always the risk of concealing negative effects that might have been recognized if the vaccine had been evaluated in bigger populations [77].
A successful vaccine for SARS-CoV-2 should be safe, effective, robust, provide long-lasting immunity, and be deployable to a large population. Currently, there are more than 50 ongoing SARS-CoV-2 vaccine clinical trials, of which majorly the vaccine strategy showing maximum protection utilizes the S protein of the virus. Though some of the studies have shown promise in the non-human primate models, still further validations are needed [78].
Vaccine design is a hard task in itself considering many challenges in the way. Figure 3 shows the major obstacles which come during the SARS-CoV-2 vaccine development.
It has been discussed above the different vaccine candidates being explored or are in phase trials currently as shown above. The National Library of Medicine of the US National Institute of Health has recorded about 80 clinical studies in the Clinical Trial Database to date. Out of these, 11 of them are in phase 1,8 of them are in phase 1/2, 3 of them in phase 2, 1 in phase 2/3, and 11 are there in phase 3 [79].
Any kind of vaccine when designed should take care of the potential unwanted side effects. During the SARS-CoV-2 pandemic’s second wave, it was observed that many affected individuals died, just after receiving the vaccines. The theories which are coming up that support this kind of exacerbated immune response is that the pre-existing antibodies to other coronaviruses triggered the immune response, and a storm of cytokines was initiated through antibody-dependent enhancement, which lead to a number of mortalities [75]. Animal studies have also shown that neutralizing antibodies to S protein can potentially exacerbate severe lung injury in SARS-CoV-2 infection by aggravating immunological responses. Also, it has been shown that there is a strong correlation between antiviral IgG seroconversion and ARDS in approximately 80% of the patients [80].
Though there is a structural similarity between SARS-CoV-2 and SARS-CoV, they share the same binding sites on the host receptors. Therefore, they also share the same pathogenesis and exhibit cross-immunity to much extent. This has helped researchers across the world to develop a vaccine against COVID-19 in the shortest of time. An interesting fact about coronaviruses s that if seen carefully, this 21st century has been so welcoming for this family of viruses. Almost 3 types of epidemics and a pandemic like SARS, MERS, COVID-19 have been experienced [81]. And similar disease spreads are also expected in the future too, so there is a need to study the most conserved and homologous molecules of such viruses. Like in CoV’s S2 subunit seems very promising for developing a Pan-CoV vaccine that can be used worldwide for vaccinating all kinds of populations.
Vaccine against SARS-CoV-2 in children
The road to developing a COVID-19 vaccine has been long and winding, but it has yielded promising results. In the United States and many other countries, the COVID-19 vaccination is currently available for children aged 12 to 15. Pfizer/BioNTech, Astra Zeneca, Moderna, and Bharat Biotecetc are some of the companies that are in phase 3 trials of their vaccines. Some of them are also out in the market, Food and Drug Administration (FDA) approved and have also been tested in children below 18 years of age. These have shown promising results with 100 percent efficacy. In India, Covaxin and Zycov-D (ZydusCadila) Vaccines are among the top priority of the Indian Government. Zycov D has completed its phase trials and has been approved by the Indian Government. It’s a needle-free vaccine and has been promised to be available for children 12–18 years of age.
The other company vaccines for children like Pfizer, Covaxin and Sputnik are also underway to complete their trials and will also be on market soon. These vaccines for children are mostly using the much safer method of vaccine preparation, where components of the virus for example spike like structure (S protein), etc. are being explored for instructing the cells especially the host immune cells, to make a piece of harmless S protein and exposing them on the cell surfaces. These proteins are thus detected by the immune system, which triggers an immunological response that leads to the production of antibodies against them. Once the protein pieces are made, proof reading occurs and the cells break down the instructions and get rid of them. The mRNA in the vaccine doesn’t enter into the nucleus of the cell where DNA is kept [82].
Conclusions
This pandemic, howsoever, doesn’t seem to end up abruptly, hence many lessons have been taught to mankind in this period. The most important is with vaccine development, which has always been a time taking and exhaustive mission, has gained a remarkable developmental speed during this outbreak. This landscaping of SARS-CoV-2 vaccine research and development is a revolution that has showcased the varied range of tested technological platforms for rapid vaccine development. These include VLP’s, peptides, nucleic acids (DNA and RNA), viral vector (replicative and non-replicative), recombinant proteins, live attenuated viruses, and inactivated viruses to name some, which with the conventional vaccines developed so far are not fully authorized. But with the prior experiences in the field of oncology, these technological bases are explored in COVID-19 vaccine development so that a successful vaccine for this pandemic comes at the earliest. This pandemic experience has taken many lives across the world, which motivates towards continuous exploration for the development of the most promising vaccine candidates till the point they can be stockpiled and are made ready for trials whenever an emergency or an outbreak arises or recur. We hope that we get a vital and key formulation for this disease prevention, which should be effective or should prevent even the severity or recurrence of this virus and its mutated versions, which are expected to come in the near times soon.
Abbreviations
| ACE2: | angiotensin-converting enzyme 2 |
| APCs: | antigen-presenting cells |
| CCL2: | C-C motif chemokine ligand 2 |
| COVID-19: | coronavirus disease-19 |
| CXCL8: | C-X-C motif chemokine ligand 8 |
| DCs: | dendritic cells |
| dsRNA: | double-strand RNA |
| GRP78: | glucose-regulated protein 78 |
| IFN: | interferon |
| IgG: | immunoglobulin G |
| IL-6: | interleukin-6 |
| IRF: | interferon regulatory factor |
| JAK/STAT: | Janus kinase/signal transducers and activators of transcription |
| MERS: | Middle East respiratory syndrome |
| mRNA: | messenger RNA |
| N: | nucleoprotein |
| NF-kB: | nuclear factor kappa B |
| NK: | natural killer |
| PRRs: | pattern recognition receptors |
| RBD: | REM sleep behavior disorder |
| RIG: | retinoic acid-inducible gene |
| RLRs: | retinoic acid-inducible gene-like receptors |
| S: | spike |
| SARS-CoV-2: | severe acute respiratory syndrome coronavirus 2 |
| ssRNA: | single-strand RNA |
| TBK1: | TANK-binding kinase 1 |
| Th1: | T-helper 1 |
| TLRs: | Toll-like receptors |
| TMPRSS2: | transmembrane serine protease 2 |
| TNF-α: | tumour necrosis factor alpha |
| VLP: | virus-like particle |
Declarations
Author contributions
AS, VS, AK, MOP wrote the article. BJO and MOP edited and concepted the manuscript. All authors contributed to manuscript revision, read and approved the submitted version.
Conflicts of interest
The authors declare that they have no conflict of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent to publication
Not applicable.
Availability of data and materials
Not applicable.
Funding
Not applicable.
Copyright
© The Author(s) 2022.