Affiliation:
1Department of Rheumatology and Immunology, The Second Hospital of Shanxi Medical University, Taiyuan 030000, Shanxi, China
2Shanxi Provincial Key Laboratory of Rheumatism Immune Microecology, Taiyuan 030000, Shanxi, China
3Key Laboratory of Cellular Physiology at Shanxi Medical University, Ministry of Education, Taiyuan 030000, Shanxi, China
†These authors contributed equally to this work and share the first authorship.
ORCID: https://orcid.org/0000-0001-8235-0482
Affiliation:
2Shanxi Provincial Key Laboratory of Rheumatism Immune Microecology, Taiyuan 030000, Shanxi, China
3Key Laboratory of Cellular Physiology at Shanxi Medical University, Ministry of Education, Taiyuan 030000, Shanxi, China
†These authors contributed equally to this work and share the first authorship.
ORCID: https://orcid.org/0000-0002-7989-4822
Affiliation:
2Shanxi Provincial Key Laboratory of Rheumatism Immune Microecology, Taiyuan 030000, Shanxi, China
3Key Laboratory of Cellular Physiology at Shanxi Medical University, Ministry of Education, Taiyuan 030000, Shanxi, China
Affiliation:
2Shanxi Provincial Key Laboratory of Rheumatism Immune Microecology, Taiyuan 030000, Shanxi, China
3Key Laboratory of Cellular Physiology at Shanxi Medical University, Ministry of Education, Taiyuan 030000, Shanxi, China
Affiliation:
2Shanxi Provincial Key Laboratory of Rheumatism Immune Microecology, Taiyuan 030000, Shanxi, China
3Key Laboratory of Cellular Physiology at Shanxi Medical University, Ministry of Education, Taiyuan 030000, Shanxi, China
Affiliation:
2Shanxi Provincial Key Laboratory of Rheumatism Immune Microecology, Taiyuan 030000, Shanxi, China
3Key Laboratory of Cellular Physiology at Shanxi Medical University, Ministry of Education, Taiyuan 030000, Shanxi, China
Affiliation:
2Shanxi Provincial Key Laboratory of Rheumatism Immune Microecology, Taiyuan 030000, Shanxi, China
3Key Laboratory of Cellular Physiology at Shanxi Medical University, Ministry of Education, Taiyuan 030000, Shanxi, China
Affiliation:
1Department of Rheumatology and Immunology, The Second Hospital of Shanxi Medical University, Taiyuan 030000, Shanxi, China
2Shanxi Provincial Key Laboratory of Rheumatism Immune Microecology, Taiyuan 030000, Shanxi, China
3Key Laboratory of Cellular Physiology at Shanxi Medical University, Ministry of Education, Taiyuan 030000, Shanxi, China
4SXMU-Tsinghua Collaborative Innovation Center for Frontier Medicine, Shanxi Medical University, Taiyuan 030001, Shanxi, China
Email: zhangshengxiao1@sxmu.edu.cn
Explor Med. 2024;5:709–719 DOI: https://doi.org/10.37349/emed.2024.00251
Received: April 17, 2024 Accepted: July 10, 2024 Published: October 25, 2024
Academic Editor: Tamer A. Gheita, Cairo University, Egypt
Background: Rheumatoid arthritis (RA) is an autoimmune joint disease with systemic manifestations. Emerging evidence implicates the gut microbiota in the pathophysiology of RA. However, the specific changes in the gut microbiota in RA patients remain poorly understood.
Methods: We conducted a comprehensive search of PubMed, EMBASE, Web of Science, Cochrane Library, MEDLINE, Wanfang, VIP, CBM, and CNKI from the time the databases were created until October, 2023. To evaluate changes in α-diversity and the abundance of certain microbiota families in RA, standardized mean difference (SMD) and 95% confidence interval (CI) calculations were made.
Results: Firstly, we evaluated the changes in α-diversity of gut microbes in patients with RA compared to healthy controls. Our analysis of 18 studies revealed a significant decrease in richness and evenness among RA patients. Importantly, the subgroup analysis suggested the decrease in α-diversity was more significant in treatment naïve patients rather than those who received anti-rheumatic medication. Additionally, in the subgroup analysis of 316 patients stratified by baseline disease activity, no significant differences in α-diversity were detected between groups.
Discussion: Our analysis further evidence of altered α-diversity and the relative abundance of specific bacteria in RA. These findings contribute to our understanding of the role of gut microbiota dysbiosis in the pathogenesis of RA. Further research is warranted to elucidate the underlying mechanisms and explore the potential therapeutic implications of targeting the gut microbiota in RA management.
Rheumatoid arthritis (RA) is a systemic chronic autoimmune joint disease affecting 0.5% to 1.0% of the global population [1]. It primarily affects joints but should be considered a syndrome that includes extra-articular manifestations, such as rheumatoid nodules, pulmonary involvement or vasculitis, and systemic complications [2]. The role of pathogenic microorganisms in the development of RA remains a topic of debate [3, 4]. Several bacteria, such as Mycoplasma fermentans, Escherichia coli, and Proteus mirabilis, have been proposed to be associated with RA morbidity. However, Picchianti-Diamanti et al. [5] did not observe phylum-level differences between RA and healthy individuals. Nonetheless, they found elevated levels of the Bacilli class, Lactobacillales order, and a decrease in Faecalibacterium prausnitzii species in untreated RA patients. Vaahtovuo et al. [6] reported a reduced relative abundance of Prevotella in the intestines of RA patients. However, a rise in Prevotella copri was observed in RA patients with new onset and no treatment [7]. Studies in Chinese patients indicated a higher prevalence of Prevotella copri in new-onset RA cases compared to healthy individuals, but a lower prevalence in established RA cases [8]. Additionally, active RA cases demonstrated increased levels of Lactobacillus [7]. Furthermore, a separate study indicated that individuals at risk for RA exhibited higher concentrations of Prevotella copri compared to their first-degree relatives. Further research is warranted to address these current knowledge gaps.
Despite the growing interest in gut microbiota and RA, limited research has comprehensively explored the underlying causes and mechanisms behind these microbial changes. Therefore, the objective of our study is to comprehensively analyze the existing evidence on the relationship between gut microbiota and RA.
The association between RA and gut microbiota was assessed using PubMed, EMBASE, Web of Science, Cochrane Library, MEDLINE, Wanfang, VIP, CBM, and CNKI databases from the establishment to October 22, 2023. Key search terms include gastrointestinal microbiome, microbiome, gastrointestinal tract, intestinal microbiome, bacteria, intestinal tract, microflora, gastrointestinal tract, rheumatoid, rheumatic disease, juvenile RA, pediatric RA, and their synonyms. The preferred reporting items for the systematic review and meta-analysis (PRISMA) report are followed by this study, which has been preregistered with PROSPERO (CRD42023413477).
All included studies performed research at the population level; the relationship between gut microbiota and RA was studied, the changes in gut microbiota indexes in RA and healthy controls (HCs) were compared, and data for analysis were available.
Inclusion criteria: 1) Human research; 2) Studies on the changes of gut microbiota in patients with RA.
Exclusion criteria: 1) Duplicate references in different databases; 2) Excluding review articles, guidelines, conference abstracts, or case reports; 3) Incomplete data or unable to obtain full text; 4) No HCs groups; 5) No available data were reported on intestinal flora α-diversity or intestinal microbial community composition.
We extracted the following data from the selected studies: essential information including first author, publication year, and country; clinical information including case numbers, genders, ages, α-diversity index in patients and healthy people, the relative abundance of intestinal microorganisms; experimental data including fecal collection method, storage method, DNA extraction technology.
The Newcastle-Ottawa Quality Scale (NOS) was used to evaluate the caliber of the included research. The appropriateness of the sampling frame, the sampling method, the determination of exposure, the presence of evidence of the absence of relevant outcomes at the study beginning, the comparability of cohorts, the process of outcome assessment, etc., were prerequisites for the NOS assessment of bias risk in case-control studies. Based on the eligibility requirements, two researchers independently evaluated the studies. Then, investigators were brought in to settle disagreements.
Analysis was performed using Stata12.0 (StataCorp, College Station, Texas, USA). Since continuous variables are used to indicate variety, the results were examined using standardized mean differences (SMDs) and their 95% confidence intervals (CIs) as measures of effectiveness. The heterogeneity among the results was evaluated according to the I² of the included effects. Fixed-effects models were employed when there was low heterogeneity between studies (I² ≤ 50%). In contrast, a random effect model was utilized if heterogeneity was present (I² > 50%). Publication bias was assessed by using funnel plots and the Egger test.
Electronic database searches yielded 2,957 articles. After the title and abstract review, 2,911 articles were excluded because they did not meet the inclusion criteria. A total of 318 full-text articles were retrieved. As a result, 298 articles were excluded because they did not meet the eligibility criteria, and a total of 18 articles were eligible (Figure 1).
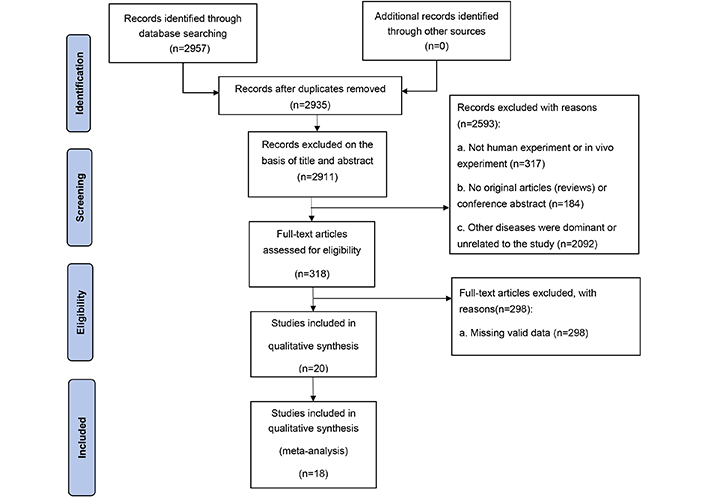
Retrieval flow chart
Note. Adapted from “The PRISMA 2020 statement: an updated guideline for reporting systematic reviews” by Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. BMJ. 2021;372:n71 (https://www.bmj.com/content/372/bmj.n71). CC BY.
Eighteen studies examined 1,166 RA patients and 968 HCs. They all used human feces as experimental samples and were observational case-control studies. According to NOS, each included study was high quality (NOS score: 6–8) (Table 1).
Basic information included in the study
| Study | Concomitant treatment | Q※ | Technology employed | Platform |
|---|---|---|---|---|
| Chen-2019 [9] | NA | 7 | 16S rRNA gene sequencing | Illumina Miseq sequencing platform |
| Liu-2012 [10] | NA | 9 | 16S rRNA gene sequencing | Illumina Miseq sequencing platform |
| Kang-2021 [11] | NA | 9 | 16S rRNA gene sequencing | Illumina Miseq sequencing platform |
| Wang-2022 [12] | NSAIDs: 100 (30/30) | 7 | ITS2-based DNA sequencing | Illumina Miseq sequencing platform |
| Yin-2021 [13] | NA | 9 | 16S rRNA gene sequencing | Illumina Miseq sequencing platform |
| Wang-2022 [14] | NSAIDs: 100 (62/62) | 9 | ITS2-based DNA sequencing | Illumina Miseq sequencing platform |
| Wang-2018 [15] | NA | 6 | NA | NA |
| Sun-2022 [16] | NA | 8 | ITS2-based DNA sequencing | Illumina Miseq sequencing platform |
| Qiao-2022 [17] | NA | 3 | ITS2-based DNA sequencing | Illumina Miseq sequencing platform |
| Breban-2017 [18] | NSAIDs: 35.3 (6/17); Corticosteroids: 82.4 (14/17); DMARDs: 53 (9/17); Biotherapy: 70.6 (12/17); Antiacid: 70.6 (12/17) | 7 | 16S rRNA gene sequencing | Illumina Miseq sequencing platform |
| Forbes-2018 [19] | NA | 9 | 16S rRNA gene sequencing | Illumina Miseq sequencing platform |
| Jeong-2019 [20] | NA | 6 | 16S rRNA gene sequencing | Illumina Miseq sequencing platform |
| Kishikawa-2020 [21] | csDMARDs: 28 (23/82); bDMARDs: 2 (2/82) | 8 | Metagenomic shotgun sequencing | Illumina high-throughput sequencing platform |
| Li-2021 [22] | NA | 9 | ITS2-based DNA sequencing | Illumina Miseq sequencing platform |
| Sun-2019 [23] | NA | 7 | ITS2-based DNA sequencing | Illumina Miseq sequencing platform |
| Wang-2022 [24] | NA | 5 | 16S rRNA gene sequencing | Illumina Miseq sequencing platform |
| Jin-2023 [25] | NA | 8 | 16S rRNA gene sequencing | Illumina Miseq sequencing platform |
| Koh-2023 [26] | NA | 8 | 16S rRNA gene sequencing | Illumina Miseq sequencing platform |
※ Quality (Q) of each study was based on the Newcastle-Ottawa Quality. NA: not applicable
Of 18 studies that investigated α-diversity differences between RA and HCs, we obtained six α-diversity indices. We included them in the meta-analysis (1,166 patients and 968 controls), including richness (ACE, Chao1, observed species), evenness, and richness/evenness (Shannon, Simpson). The most widely used were Shannon, Simpson, ACE, and Chao1.
Concerning diversity, a significant decrease in the Shannon index was observed among RA patients compared to HCs, as reported in fifteen studies (Figure 2A: SMD = –0.509, 95% CI: –0.798, –0.220; P < 0.05, I2 = 87.9%). Additionally, the Simpson index showed a significant decrease among RA patients compared to HCs (Figure 2B: SMD = –0.467; 95% CI: –0.788, –0.145; P = 0.004). Regarding richness, data from nine studies demonstrated a significant decrease in ACE among RA patients compared to HCs (Figure 2C: SMD = –0.330, 95% CI: –0.622, –0.039; P = 0.026, I2 = 83.5%). Similarly, data from eleven studies indicated a significant decrease in Chao1 among RA patients compared to HCs (Figure 2D: SMD = –1.213; 95% CI: –1.770, –0.656; P < 0.001, I2 = 96.0%). Regarding evenness, a significant difference was observed between RA patients and HCs based on data from two studies (Figure 2E: SMD = –1.863, 95% CI: –3.181, –0.544; P = 0.006, I2 = 88.0%). However, there was no significant difference in observed species between the two groups, as evidenced by data from two studies (Figure 2F: SMD = –4.342, 95% CI: –9.098, 0.415; P = 0.074).
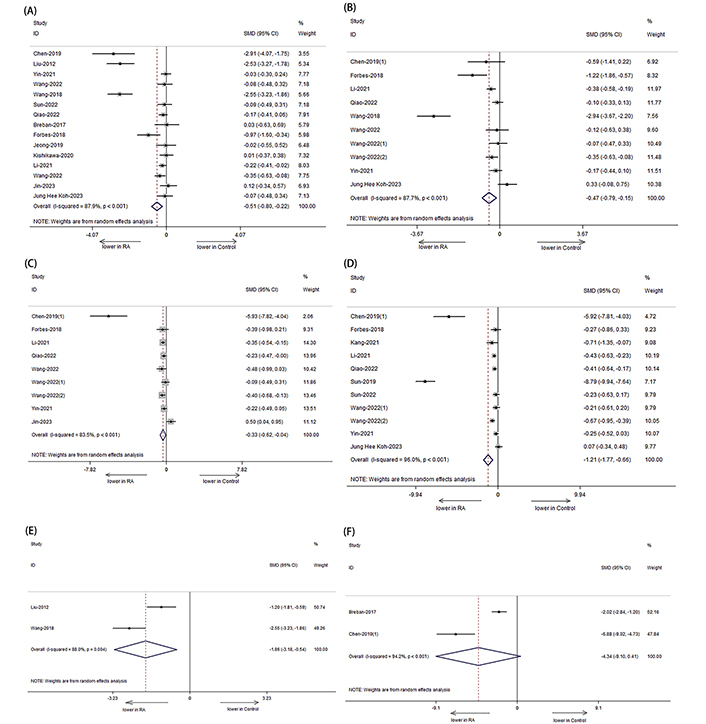
Forest plots of alterations in the α-diversity of patients with RA versus HCs. (A) Shannon index; (B) Simpson index; (C) ACE; (D) Chao1; (E) evenness index; (F) observed species. Compared to HCs, the Shannon index, Simpson index, ACE, Chao1, and evenness index of RA patients showed a significant decrease
These results indicate a decrease in α-diversity, including reductions in richness and evenness, among patients with RA compared to HCs.
In our study, we categorized patients into two groups based on their medication usage: an on-treatment group (patients receiving anti-rheumatic medication) and a treatment naïve group (patients not receiving initial treatment). We observed that the α-diversity was significantly reduced in RA patients, particularly in the treatment naïve group (Figure 3A: SMD = –0.653; 95% CI: –1.0001, –0.305; P < 0.001; Figure 3B: SMD = –0.573; 95% CI: –0.958, –0.187; P < 0.001; Figure 3C: SMD = –0.365; 95% CI: –0.722, –0.008; P < 0.001; Figure 3D: SMD = –1.346, 95% CI: –1.956, –0.736; P < 0.001), rather than those who received disease-modifying antirheumatic drugs (DMARDs) (Figure 3A: SMD = –0.024; 95% CI: –0.276, 0.228; P > 0.05; Figure 3B: SMD = –0.093; 95% CI: –0.407, 0.220; P > 0.05; Figure 3C: SMD = –0.253; 95% CI: –0.632, 0.126; P > 0.05; Figure 3D: SMD = –0.206, 95% CI: –0.608, 0.195; P > 0.05).
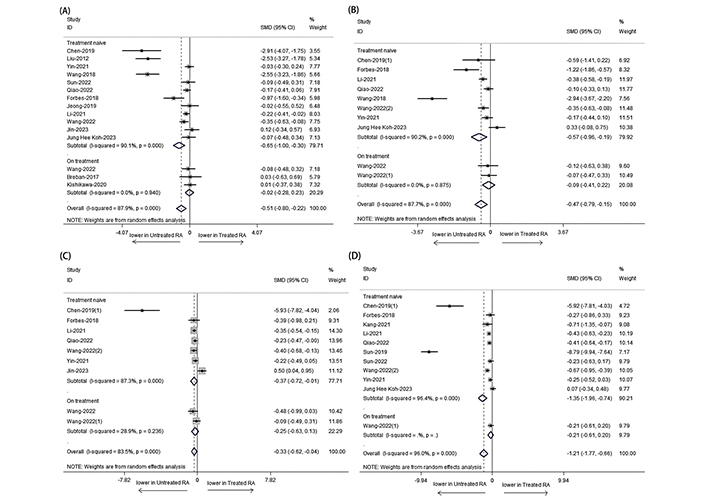
Forest plots of α-diversity for RA patients receiving anti-rheumatic medication treatment or not. (A) Shannon index; (B) Simpson index; (C) ACE; (D) Chao1. The α-diversity of RA who have not received anti rheumatic drug treatment is significantly reduced. For figure (D), the I-squared and P values not provided are not applicable. Additionally, P = 0.000 represents P < 0.001
We specifically analyzed the composition of intestinal microbial communities in patients with RA. The results showed that the RA had an increased abundance of Proteobacteria (Figure 4A: SMD = 0.806, 95% CI: 0.607, 1.006; P < 0.001), Actinobacteria (Figure 4B: SMD = 0.337, 95% CI: 0.153, 0.521; P < 0.001) but a decreased abundance of Bacteroides fragilis (Figure 4C: SMD = –0.205, 95% CI: –0.545, –0.136; P < 0.001) compared with those of HCs (Figure 4).
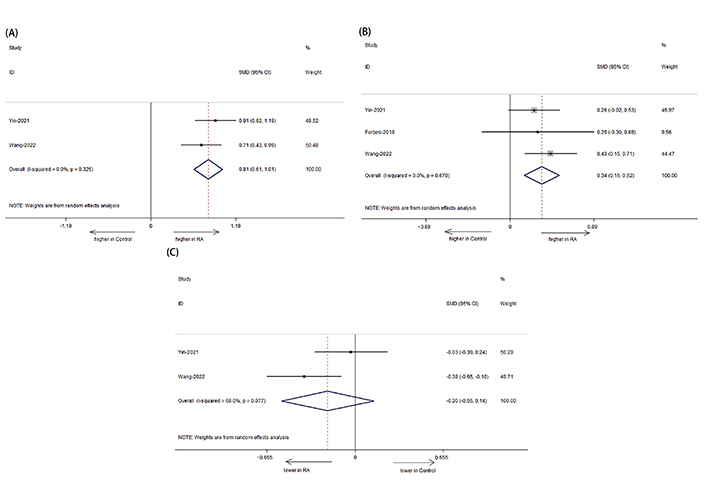
Forest plots of alterations in the gut microbiota of patients with RA versus HCs. (A) Proteobacteria; (B) Actinobacteria; (C) Bacteroides fragilis. Compared to HCs, RA had an increased abundance of Actinobacteria and Proteobacteria, but a decreased abundance of Bacteroides fragilis
We used Egger, Begg, and funnel plots to test and assess publication bias risk. The results showed no evidence of publication bias, indicating that the conclusions of the meta-analysis were relatively reliable.
The dysregulation of the gut microbiota is believed to be a contributing factor in the development of RA [27]. However, the role of gut microbiota in immunological tolerance among RA patients remains unclear. Previous studies have reported conflicting results concerning changes in gut microbiota diversity in RA patients, leading to an ongoing debate regarding the gut microbiota status in this patient population. To gain a better understanding of the role of gut microbiota in RA, we conducted a meta-analysis examining alterations in the gut microbiota of RA patients.
In our study, we observed significant differences in α-diversity between patients with RA and HCs, which aligns with findings reported in previous studies [28]. It is noteworthy that alterations in the composition and diversity of the gut microbiota can impact immune regulation via the lymphatic or circulatory systems [29]. During periods of reduced immunity, pathogenic microorganisms can stimulate the dominant microbiota within the gut, leading to the secretion of various inflammatory mediators that may promote the onset of RA.
Our results demonstrated a significant difference in the richness of the gut microbiota in RA, particularly in the abundance of Proteobacteria, Actinobacteria, and Bacteroides fragilis. Specifically, Proteobacteria, a Gram-negative bacterium, showed a significant increase in the RA groups. This bacterium primarily activates the innate immune system through lipopolysaccharide (LPS), and LPS-binding protein (LBP) plays a crucial role in rapidly inducing the inflammatory response to small amounts of LPS or Gram-negative bacteria. LBP binds to LPS and transfers it to the CD14 LPS receptor on macrophages or high-density lipoprotein particles [30]. Additionally, Actinobacteria showed an increased presence in RA, and its abundance was found to be a potential predictor of RA status. Another noteworthy gut microbiota, Bacteroides fragilis, was found to induce the secretion of IL-10 by CD4+ T cells and is closely associated with the immune profile of RA [31]. Our findings provide valuable complementary data to existing research in this field. Furthermore, we did not observe any statistically significant differences between the RA and HCs in terms of Firmicutes, Bacteroidetes, Bifidobacterium, Lactobacillus, Enterobacterium, Enterococcus, and Clostridium. Although these microbiotas have been previously associated with RA, it is important to consider the variations in dietary habits among different races, cultures, and regions. Thus, the results of other studies may not be directly applicable to our study due to these differences in eating habits.
The dysbiosis of the gut microbiota is an intrinsic attribute of RA but not due to the treatment. We observed a significant decrease in α-diversity in patients who did not receive initial treatment. Treatment with DMARDs can have a significant impact on the gut microbiota. Previous studies have demonstrated that methotrexate inhibits aminoimidazole carboxamide ribonucleotide (AICAR) transformylase, leading to increased intracellular AICAR levels and subsequent accumulation and release of adenosine. This activation of A2A and A3 adenosine receptors results in anti-inflammatory properties [32]. Additionally, the microbial metabolism of sulfasalazine produces active anti-inflammatory products [33]. Furthermore, the homeostasis of the gut microbiota plays a crucial role in the immune system and can modulate the intestinal metabolism of drugs. Any abnormal changes in the gut microbiota can lead to altered drug response [34].
On the other hand, another study suggested that dysbiosis of gut microbiota may play a role in disease activity and treatment outcomes [35]. The gut microbiome plays an important role in shaping the immune system and promoting the pathogenesis of many diseases. Vitamin D is crucial for maintaining a healthy microbiome and is involved in regulating immune responses [36]. Studies have shown that vitamin D deficiency is associated with dysregulation of the microbiome, leading to an increase in Proteobacteria and inflammatory diseases [37]. The metabolites of Proteobacteria will stimulate the differentiation and development of the Th17 and Treg cells, which may lead to excessive or inhibitory immune response. Meanwhile, vitamin D promotes T cell differentiation and function, which is also associated with chronic inflammation. Murdaca et al. [38] discussed a wealth of evidence, confirming that the immune system and microbiome are interrelated, and vitamin D is a key intermediate participant in this dynamic. The microbiome and vitamin D affect each other and the immune system in various ways. Moreover, the use of shotgun metagenomic sequencing of microbial communities in stool samples has shown promise as a reliable predictor of whether patients with RA will experience significant improvement in disease activity [39]. However, in our study, we did not observe a correlation between disease activity and gut microbiota.
Our research is subject to certain limitations that should be acknowledged. Firstly, there is considerable heterogeneity across studies, which can be attributed to variations in diagnostic criteria for RA, gender distribution, and medication usage among patients [20]. These factors introduce potential sources of variability and may impact the generalizability of our findings. Secondly, the credibility of our conclusions may be questioned due to limited data availability on factors such as uniformity of species observed and specific phyla. Thirdly, as there was no data available on disease activity in the study database, our study may suffer from residual confounding by disease activity if there is a strong association between disease activity and gut microbiota. To ensure the reliability of our investigation, we excluded studies of low quality. However, even when we restricted our analysis to studies with a low risk of bias, the estimates did not considerably change. Nonetheless, future research on related topics is warranted to generate more robust evidence-based medical knowledge. While our study has its limitations, including study heterogeneity and limited data on certain important factors, we have taken steps to address these limitations and present reliable findings. To strengthen the existing evidence base, further research should be conducted to explore related aspects of this subject matter.
Overall, our study revealed significant differences in the gut microbiota between RA and HCs. The decrease in α-diversity expression was more pronounced in untreated patients than in patients treated with anti-inflammatory drugs. However, the relationship between gut microbiota and disease activity remains unclear. Further research is needed to clarify the effects of baseline RA disease activity on the α-diversity of gut microbiota. Further research addressing study heterogeneity and exploring specific microbial species and phyla is necessary to enhance our understanding of the intricate relationship between the gut microbiota and RA, potentially leading to novel therapeutic interventions.
CI: confidence interval
HCs: healthy controls
LPS: lipopolysaccharide
RA: rheumatoid arthritis
SMDs: standardized mean differences
We would like to thank all the participants involved in this study.
QYS: Conceptualization, Investigation, Writing—review & editing. YZ: Data curation. DQ: Writing—original draft. XS and YS: Investigation, Data curation. RQL and SXZ: Data curation, Formal analysis, Validation. YFZ: Data curation. All the authors read and approved the final manuscript.
The authors declare that they have no conflicts of interest.
Not appliable.
Not appliable.
Not appliable.
The original data supporting the conclusions of this manuscript will be made available free of charge to any qualified researcher by the author, Sheng-Xiao Zhang, if required.
This study was supported by the Natural Science Foundation of Shanxi Province No. [202203021221269]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
© The Author(s) 2024.
Copyright: © The Author(s) 2024. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
