Affiliation:
Laboratory of Medical Statistics and Biometry, “Giulio A. Maccacaro”, Department of Clinical Sciences and Community Health, University of Milan, 20133 Milan, Italy
Email: danila.coradini@gmail.com
ORCID: https://orcid.org/0000-0003-3771-4341
Explor Med. 2024;5:950–959 DOI: https://doi.org/10.37349/emed.2024.00268
Received: September 10, 2024 Accepted: November 15, 2024 Published: December 15, 2024
Academic Editor: Andrea Nicolini, University of Pisa, Italy
The article belongs to the special issue Breast Cancer: Basic and Clinical Advances
Aim: Using a dataset available from the NCBI Gene Expression Omnibus Repository, this in silico study investigated the differential expression of GPX4, the gene coding for the detoxifying enzyme glutathione peroxidase paired samples of AH (atypical hyperplasia) and corresponding histologically normal (HN) tissue from 17 women with AH and in four samples of normal breast tissue used as controls.
Methods: The study focused on the genes (HMGCR, FDPS, FDFT1, and GGPS1) involved in the production and breakdown of isopentenyl-diphosphate, a key component for GPX4 biosynthesis. It also explored the connection between the expression of GPX4 and the genes (CCND1, CDK4, CDK6, and CDKN1B) associated with the cyclin D1-CDK4/6 complex.
Results: Compared to HN tissue, AHs exhibited higher levels of GPX4 and HMGCR, supporting the functional connection between GPX4 synthesis and isopentenyl-diphosphate production. Additionally, AHs showed elevated levels of CCND1 and CDKN1B and decreased levels of CDK6. Compared to normal breast tissue, HNs showed similar alterations, suggesting that ferroptosis escape and uncontrolled proliferation are early molecular events in the neoplastic transformation. Compared to HN tissue, AHs also expressed high levels of GGPS1, a downstream gene of HMGCR, which leads to the synthesis of geranylgeranyl-diphosphate, a molecule essential for the post-translational modification of the proteins involved in the regulation of the Hippo signaling pathway.
Conclusions: Although very preliminary, present results seem to suggest that blocking the mevalonate pathway by statins might, on the one hand, prevent AHs from escaping ferroptosis through depleting isopentenyl-diphosphate and, on the other hand, inhibit cell proliferation by controlling the Hippo pathway.
The term “atypical hyperplasia” (AH) of the breast refers to abnormal epithelial proliferative lesions that are not qualitatively or quantitatively abnormal enough to be classified as carcinoma in situ. Like most breast cancer precursors, AHs develop in the terminal duct lobular unit (TDLU), the basic functional structure of the mammary gland consisting of a bilayered ductal system ending in small glandular structures called acini [1], and are categorized into atypical ductal hyperplasia (ADH) and atypical lobular hyperplasia (ALH) based on their morphologic characteristics [2]. ADH consists of a proliferating epithelial cell population of luminal origin that strongly expresses the estrogen receptor (ER) [3]. ALH instead consists of small, round, uniform, and proliferating epithelial cells derived from the acinar population of the TDLU. These cells have an increased nucleus/cytoplasmic ratio and are E-cadherin negative [4].
Typically, AHs are found incidentally during routine screening mammography. Epidemiological evidence indicates that about 10% of breast biopsies reveal an AH and that patients with an AH at biopsy have a lifetime risk of 15%–20% of developing breast cancer [5, 6]. The standard care to prevent the progression of the AH toward invasive malignancies is surgical excision [7].
Molecular evidence has suggested that the increased proliferative activity of AHs and their progression to invasive cancer are mainly due to a failure in the balanced mechanism that controls cell proliferation and cell death [8]. However, the early events that trigger and promote the transformation of normal epithelial cells into hyperplasic cells are still unknown.
Accumulating evidence suggests that the long-term persistence in the breast tissue of some highly reactive oxygen species (ROS), reacting spontaneously with many cellular components, could play a crucial role in the transformation process [9]. In particular, the accumulation of lipid peroxides can cause extensive and irreversible damage to cell membranes and trigger ferroptosis, a form of non-apoptotic death [10].
Aimed at neutralizing the dangerous effects of ROS, preventing lipid peroxidation, and hampering ferroptosis activation, cells have developed a defense mechanism based on the detoxifying properties of glutathione peroxidases (GPX). In particular, GPX4, a selenocysteine-containing GPX, is the only member in this family of enzymes that can reduce complex lipid hydroperoxides even when embedded in membranes, thus preventing membrane disruption and ferroptosis activation [11, 12].
The biosynthesis of GPX4 requires the presence of the isopentenyl-diphosphate, an intermediate of the mevalonate pathway, the core of the biosynthetic process leading to cholesterol (Figure 1), for transferring the unusual amino acid selenocysteine to the nascent catalytic site of GPX4 [13].
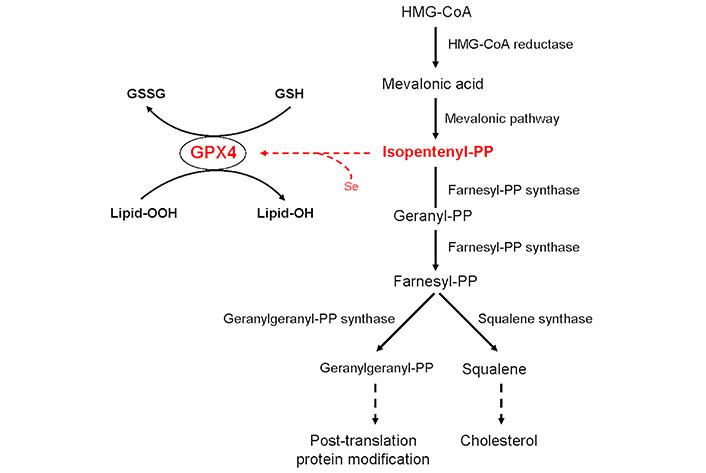
Schematic description of the relationship between glutathione peroxidase 4 (GPX4) and isopentenyl-diphosphate (isopentenyl-PP) synthesis and metabolism. Farnesyl-PP: farnesyl-diphosphate; Geranyl-PP: geranyl-diphosphate; Geranylgeranyl-PP: geranylgeranyl-diphosphate; GSH: glutathione; GSSG: glutathione disulfide; HMG-CoA: 3-hydroxy-3-methylglutaryl coenzyme-A; Lipid-OH: lipid alcohol; Lipid-OOH: lipid peroxide; Se: selenium. Solid arrows indicate the sequence of the enzymatic reactions whereas dashed arrows indicate the final biological event
Since the emerging evidence suggests that tumor cells escape ferroptosis by producing high levels of the GPX4 enzyme, thus contributing to cell growth promotion, acquired drug resistance, and cancer progression [14–16], this in silico study aimed to investigate if, like invasive cancer, AHs overexpressed the GPX4 gene when compared to the corresponding histologically normal (HN) tissue, and explore the relationship between the expression of GPX4 and that of the genes involved in the production and breakdown of isopentenyl-diphosphate (namely, HMGCR, FDPS, GGPS1, and FDFT1). The study also investigated the relation between the expression of GPX4 and that of genes coding for the elements of the cyclin D1-CDK4/6 complex, which controls the G1 phase of the cell cycle. Considering the diverse origins (ductal versus lobular portion of the TDLU) and a different risk of developing invasive cancer, ADH and ALH were analyzed separately.
The study utilized a dataset publicly available from the NCBI Gene Expression Omnibus (GEO) Repository (https://www.ncbi.nlm.nih.gov/geo/) identified by the GEO accession number GSE118432. As described in the original article [17], the study, which was approved by the Institutional Review Board of Baystate Health, Springfield, MA, under protocol number 182463, focused on women diagnosed with AH and with no prior history of breast cancer. Seventeen women with isolated AH on primary excisional biopsies and subsequent reduction mammoplasty participated in the study. Paired samples of AH and corresponding HN tissue were collected with informed consent from the patients. The HN tissue was collected at least 1 cm away from AH in the same formalin-fixed and paraffin-embedded tissue block or a different block. The case series comprised eight lobular lesions (ALH and/or classic lobular carcinoma in situ) and nine ductal lesions (ADH and/or flat epithelial atypia). Four independent samples of normal breast tissue (NOR) were collected to serve as a control. The complete transcriptome of the samples was evaluated using the [HuGene-1_0-st] Affymetrix Human Gene 1.0 ST Array [transcript (gene) version] (GEO accession GPL6244), and the expression estimates, which were filtered and log2 transformed, were uploaded in the GEO repository.
In addition to GPX4, which codes for glutathione peroxidase 4, eight genes were selected for the study. Of these genes, four code for crucial enzymes involved in isopentenyl-diphosphate synthesis and metabolism (Figure 1): HMGCR (3-hydroxy-3-methylglutaryl coenzyme-A reductase), which encodes the enzyme that converts HMG-CoA into mevalonic acid; FDPS (farnesyl-diphosphate synthase), which encodes the enzyme that catalyzes the synthesis of the farnesyl-diphosphate with geranyl-diphosphate as an intermediate step; FDFT1 (farnesyl-diphosphate farnesyltransferase 1), which encodes the enzyme that catalyzes the formation of squalene, the committed intermediate in the biosynthesis of cholesterol and sterols; GGPS1 (geranylgeranyl-diphosphate synthase 1), which encodes the enzyme that converts geranyl-diphosphate to geranylgeranyl-diphosphate, an essential element for the post-translational geranylgeranylation of the proteins. The other four genes code for the components of the cyclin D1-CDK4/6 complex: CCND1 (cyclin D1), CDK4 (cyclin-dependent kinase 4), CDK6 (cyclin-dependent kinase 6), and CDKN1B (cyclin-dependent kinase inhibitor 1B) [18].
Shapiro-Wilk was used to test normal distribution. The non-normal distribution was expressed with median and interquartile range (IQR), and non-parametric tests were used. Wilcoxon test was used for paired samples. For non-paired samples, Kruskal-Wallis and Wilcoxon tests were used. Spearman correlation coefficient was used to evaluate the correlation between genes. R Core Team version 4.1.2 (http://www.R-project.org) was used to analyze. P < 0.05 was defined as statistically significant.
Kruskal-Wallis test indicated that AHs expressed a significantly higher level of GPX4 than the corresponding HN tissue, which in turn expressed a statistically significant higher level of GPX4 than normal tissue (Figure 2A). In particular, the Wilcoxon test indicated that all the comparisons were statistically significant (HN versus NOR: 0.94 vs. 0.73, P = 0.0176; AH versus HN: 1.15 vs. 0.94, P = 0.0229; AH versus NOR: 1.15 vs. 0.73, P = 0.0063).
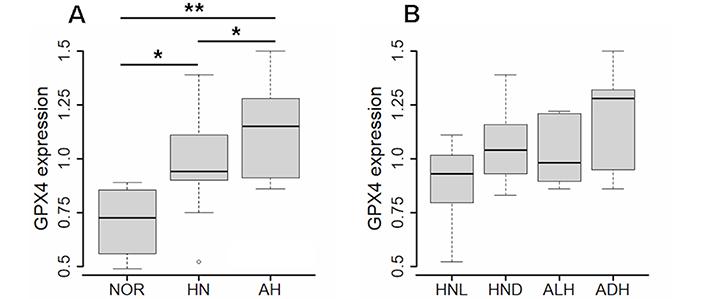
Expression of GPX4 gene in normal tissue, histologically normal tissue and atypical hyperplasia. (A) Expression of GPX4 in atypical hyperplasia (AH, N = 17), corresponding histologically normal (HN, N = 17) tissue, and normal breast tissue (NOR, N = 4). (B) Expression of GPX4 in atypical lobular hyperplasia (ALH, N = 8), atypical ductal hyperplasia (ADH, N = 9), and corresponding HN tissue [respectively, HNL (HN lobular), N =8, and HND (HN ductal), N = 9]. The differences were evaluated by the unpaired Wilcoxon test; values of P < 0.05 were considered statistically significant, and marked as * (P < 0.05) and ** (P < 0.01)
When ALH and ADH were analyzed individually, the level of GPX4 in the two subtypes was not statistically different. Likewise, the level of GPX4 did not differ in the HN tissue adjacent to the two AH subtypes (Figure 2B).
Kruskal-Wallis test indicated that AHs expressed a level of CCND1 higher than the corresponding HN tissue or NOR (Figure 3A) as confirmed by the Wilcoxon test, according to which AH versus HN: 0.96 vs. 0.76, P = 0.0099, and AH versus NOR: 0.96 vs. 0.75, P = 0.0011.
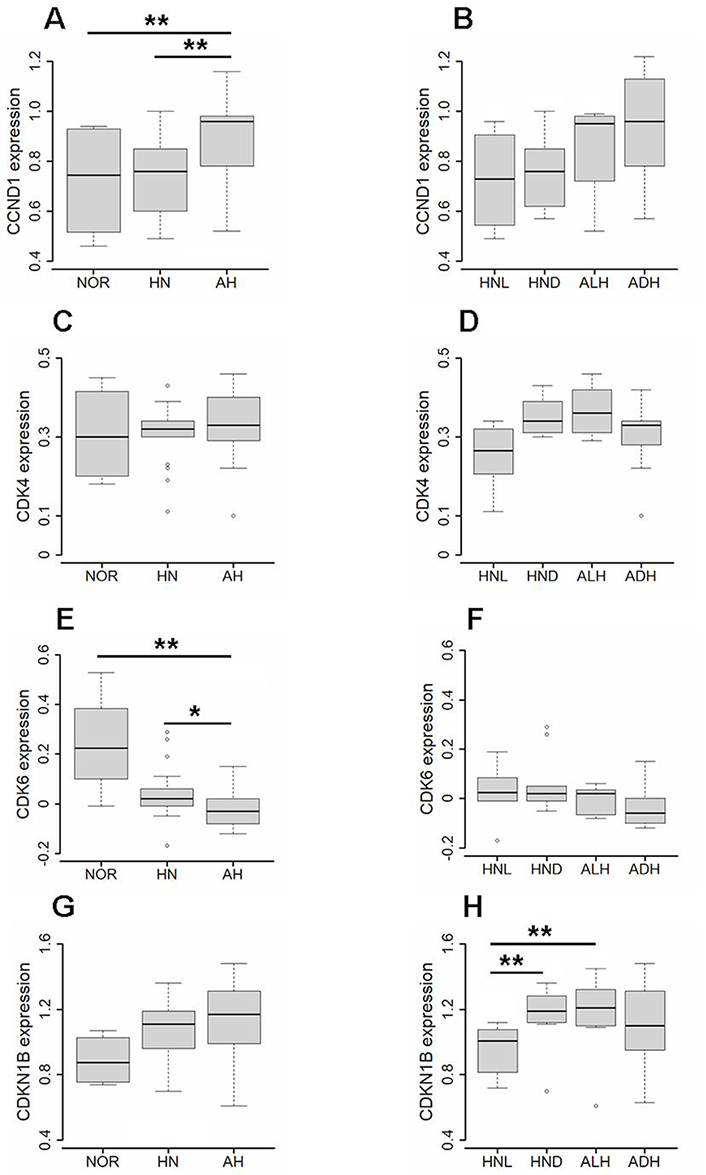
Expression of CCND1, CDK4, CDK6, and CDKN1B gene in normal tissue, histologically normal tissue and atypical hyperplasia. (A) Expression of CCND1 in atypical hyperplasia (AH, N = 17), corresponding histologically normal (HN, N = 17) tissue, and normal breast tissue (NOR, N = 4) evaluated by the unpaired Wilcoxon test. (B) Expression of CCND1 in atypical lobular hyperplasia (ALH, N = 8), atypical ductal hyperplasia (ADH, N = 9), and corresponding HN tissue (respectively, HNL, N = 8, and HND, N = 9) evaluated by the paired Wilcoxon test. (C) Expression of CDK4 in AH, HN, and NOR subgroups evaluated by the unpaired Wilcoxon test. (D) Expression of CDK4 in ALH, ADH, HNL, and HND subgroups evaluated by the paired Wilcoxon test. (E) Expression of CDK6 in AH, HN, and NOR subgroups evaluated by the unpaired Wilcoxon test. (F) Expression of CDK6 in ALH, ADH, HNL, and HND subgroups evaluated by the paired Wilcoxon test. (G) Expression of CDKN1B in AH, HN, and NOR subgroups evaluated by the unpaired Wilcoxon test. (H) Expression of CDKN1B in ALH, ADH, HNL, and HND subgroups evaluated by the paired Wilcoxon test. Values of P < 0.05 were considered statistically significant and marked as * (P < 0.05) and ** (P < 0.01)
When ALH and ADH were considered individually (Figure 3B), the expression level of CCND1 was similar in the two AH subtypes (0.95 and 0.96, respectively) and higher than the corresponding HN tissue even not in a statistically significant manner [ALH versus HNL (HN lobular): 0.95 vs. 0.73, P = 0.0584; ADH versus HND (HN ductal): 0.96 vs. 0.76, P = 0.0742].
As regards the other components of the cyclin D1-CDK4/6 complex, while the expression of CDK4 was similar in all tissues (Figure 3C), also when ALH and ADH were considered individually (Figure 3D), the expression of CDK6 significantly decreased in AHs and corresponding HN tissue when compared to NOR (Figure 3E) as confirmed by the Wilcoxon test according to which AH versus HN: –0.03 vs. 0.02, P = 0.0221, and AH versus NOR: –0.03 vs. 0.23, P = 0.0089. The expression of CDKN1B, on the contrary, showed an increasing trend in AHs and corresponding HN tissue when compared to NOR (Figure 3G), although the difference did not reach statistical significance (AH versus NOR: 1.17 vs. 0.88, P = 0.0648, and HN versus NOR: 1.11 vs. 0.88, P = 0.0911).
When ALH and ADH were analyzed individually, CDK6 was found expressed similarly in the two AH subtypes and their corresponding HN tissue (Figure 3F), whereas CDKN1B was significantly less expressed in the HN surrounding ALH than the HN surrounding ADH (HNL versus HND: 1.01 vs. 1.19, P = 0.0093) or ALH (HNL versus ALH: 1.01 vs. 1.21, P = 0.0078) (Figure 3H).
Correlation analysis indicated that in addition to the differential expression found in AH compared to the corresponding HN or NOR, CCND1, CDK6, and CDKN1B genes significantly changed their interrelationship according to the tissue. While in NOR, CCND1 was highly associated with both genes (CCND1*CDK6: r = 0.80 and CCND1*CDKN1B: r = 0.81), in HN tissue, the association profile changed depending on the AH subtype. Thus, the HN tissue adjacent to ALH showed a correlation degree between CCND1 and CDK6 similar to that of the NOR (r = 0.80), which substantially decreased in ALH (r = 0.49); in the HN tissue adjacent to ADH, the degree of correlation between the two genes decreased dramatically (r = 0.33) and disappeared in ADH (r = 0.10). Furthermore, the positive association of CCND1 with CDKN1B decreased significantly in both AH subtypes (ALH: r = 0.49 and ADH: r = 0.57) and corresponding HN tissue (HNL: r = 0.38 and HND: r = 0.41). Conversely, the correlation analysis showed a substantial increase in the degree of association between CDKN1B and GPX4 in both AH subtypes (ALH: r = 0.81 and ADH: r = 0.78) when compared to the corresponding HN tissue (HNL: r = 0.45 and HND: r = 0.44).
Statistical analysis indicated that, of the genes involved in the production and metabolism of isopentenyl-diphosphate, only GGPS1 was differentially expressed in AHs compared to the corresponding HN tissue (AH versus HN: 0.77 vs. 0.61, P = 0.0141) and that, when ALH and ADH were considered individually, the ADH subtype expressed a higher level of GGPS1 than ALH (ADH versus ALH: 0.82 vs. 0.62, P = 0.0236) (Figure 4).
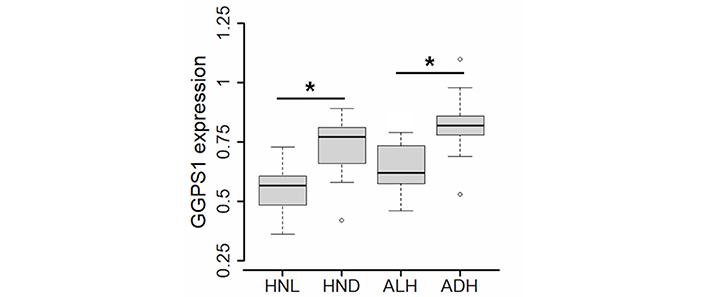
Expression of GGPS1 gene in atypical lobular hyperplasia (ALH, N = 8), atypical ductal hyperplasia (ADH, N = 9), and corresponding HN tissue (respectively, HNL, N = 8, and HND, N = 9) evaluated by the paired Wilcoxon test. Values of P < 0.05 were considered statistically significant and marked as * (P < 0.05)
Likewise, the HN tissue surrounding ADH expressed a higher level of GGPS1 than the HN tissue surrounding ALH (HND versus HNL: 0.77 vs. 0.57, P = 0.0274). Correlation analysis (Figure 5) showed that also the association of GPX4 with GGPS1 increased significantly in ADH and the corresponding HND tissue (respectively, r = 0.75, P = 0.0205 and r = 0.72, P = 0.0369) whereas, in ALH and corresponding HNL tissue, the association of the two genes was (respectively, r = 0.37, P = 0.3652 and r = 0.55, P = 0.1710) similar to that found in NOR (r = 0.40, P = 0.2912).
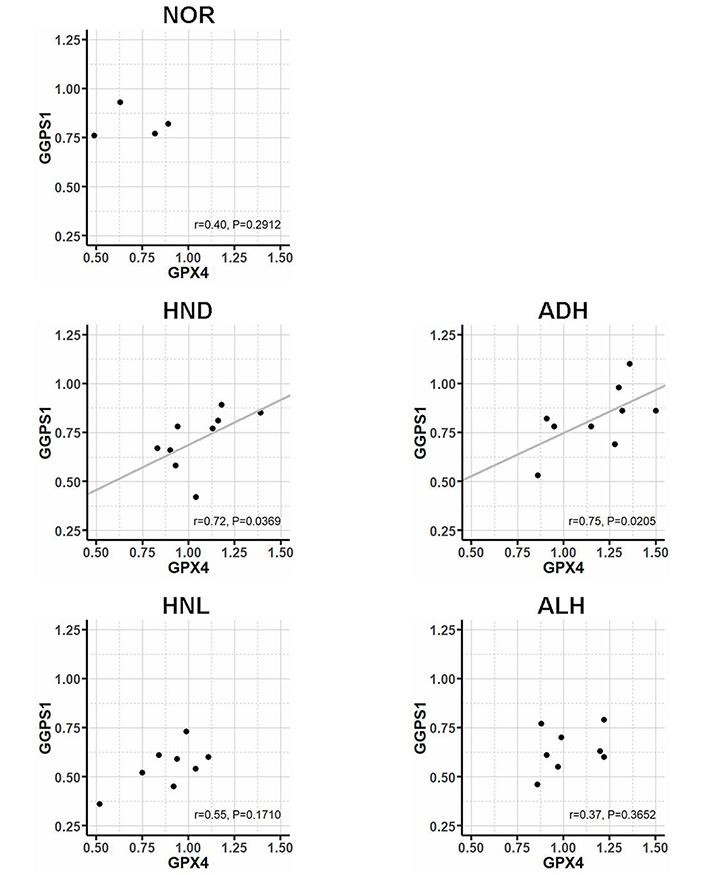
Correlation between the expression of GPX4 and GGPS1 gene in normal breast tissue (NOR, N =4), atypical ductal hyperplasia (ADH, N = 9), atypical lobular hyperplasia (ALH, N = 8), and corresponding HN tissue (respectively, HNL, N = 8, and HND, N = 9) evaluated by Spearman correlation coefficient
The finding that AHs expressed a level of the GPX4 gene higher than the surrounding HN tissue suggests that they could exploit the detoxifying property of this enzyme to escape ferroptosis. Furthermore, the observation that the HN tissue expressed a higher level of GPX4 compared to NOR indicates that the overexpression of GPX4 is an early molecular event that may predispose the normal tissue to transform into AH and proliferate. The differential expression of the genes that control the G1 phase of the cell cycle (specifically, the increased expression of CCND1 and CDKN1B and the substantial decrease in the expression of CDK6) along with the significant increase in the association between CDKN1B and GPX4, further supports the link between ferroptosis escape and cell proliferation.
As regards the association between the mevalonate pathway and GPX4 biosynthesis, present findings indicated that AHs expressed high levels of HMGCR compared to the corresponding HN tissue, in line with the augmented requirement of isopentenyl-diphosphate because of GPX4 overexpression. Remarkable is also the finding that AHs overexpressed GGPS1, which encodes for the enzyme that synthesizes geranylgeranyl-diphosphate, a molecule playing an essential role in the posttranslational prenylation of several proteins, including Rho GTPases, which act as molecular switches in various cell processes and signaling pathways, including the Hippo signaling pathway [19]. By controlling the nuclear translocation of the Yes-associated protein (YAP) coactivator, the Hippo signaling pathway regulates the expression of several genes involved in cell proliferation promotion and apoptosis inhibition [20, 21]. The observation that ADHs, the most aggressive of the two AH subtypes, expressed higher levels of GGPS1 compared to ALHs, along with the finding that in ADHs and the corresponding HN tissue, the expression of GGPS1 associated with that of GPX4 suggests that the propensity of ADH to proliferate and transform into invasive cancer could be the result of the concomitant escape from ferroptosis, inhibition of the Hippo pathway, YAP activation, and cell proliferation.
In this complex scenario, the mevalonate pathway should play a crucial role in providing the isopentenyl-diphosphate required for the biosynthesis of GPX4 and the geranylgeranyl-diphosphate necessary for Rho GTPases prenylation.
Although the findings are based on a small case series and lack support from immunohistochemical evaluation of the corresponding proteins, current results suggest that using statins to inhibit HMG-CoA reductase could be a safe and effective approach to prevent the progression of AHs (especially ADHs) to invasive cancer, resulting in the concurrent promotion of ferroptosis and inhibition of Rho GTPase prenylation. Anyway, further clinical studies are needed to confirm this promising therapeutic approach.
ADH: atypical ductal hyperplasia
AH: atypical hyperplasia
ALH: atypical lobular hyperplasia
CCND1: cyclin D1
CDK: cyclin-dependent kinase
CDKN1B: cyclin-dependent kinase inhibitor 1B
FDFT1: farnesyl-diphosphate farnesyltransferase 1
FDPS: farnesyl-diphosphate synthase
GEO: Gene Expression Omnibus
GGPS1: geranylgeranyl-diphosphate synthase 1
GPX: glutathione peroxidases
HMGCR: 3-hydroxy-3-methylglutaryl coenzyme-A reductase
HN: histologically normal
HND: histologically normal ductal
HNL: histologically normal lobular
NOR: normal breast tissue
ROS: reactive oxygen species
TDLU: terminal duct lobular unit
YAP: Yes-associated protein
DC: Conceptualization, Investigation, Writing—original draft, Writing—review & editing.
The author declares that she has no conflicts of interest.
The study utilized summary statistics from the publicly available dataset, the NCBI Gene Expression Omnibus (GEO) Repository. The original study was approved by the Institutional Review Board of Baystate Health, Springfield, MA, under protocol number 182463. As such, this research did not involve direct contact with human participants or the collection of new personal data, and therefore, ethical approval was not required.
This study analyzed secondary data from the NCBI Gene Expression Omnibus (GEO) Repository. Therefore, consent to participate is not required.
Not applicable.
The datasets analyzed for this study can be found in the NCBI Gene Expression Omnibus (GEO) Repository (https://www.ncbi.nlm.nih.gov/geo/).
Not applicable.
© The Author(s) 2024.
Copyright: © The Author(s) 2024. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
View: 2212
Download: 29
Times Cited: 0
Ali Abdul Hussein S. AL-Janabi ... Abdul Razzak Kalaf Hassan
Danila Coradini, Federico Ambrogi
Banashree Bondhopadhyay ... Vishakha Kasherwal
Nadia Islam, Suneela Vegunta
Spoorthi Marada ... Yi Lu
Remo Poto ... Gilda Varricchi
Kaoutar Anouar Tadlaoui ... Moulay Mustapha Ennaji
